FIGURE 4.
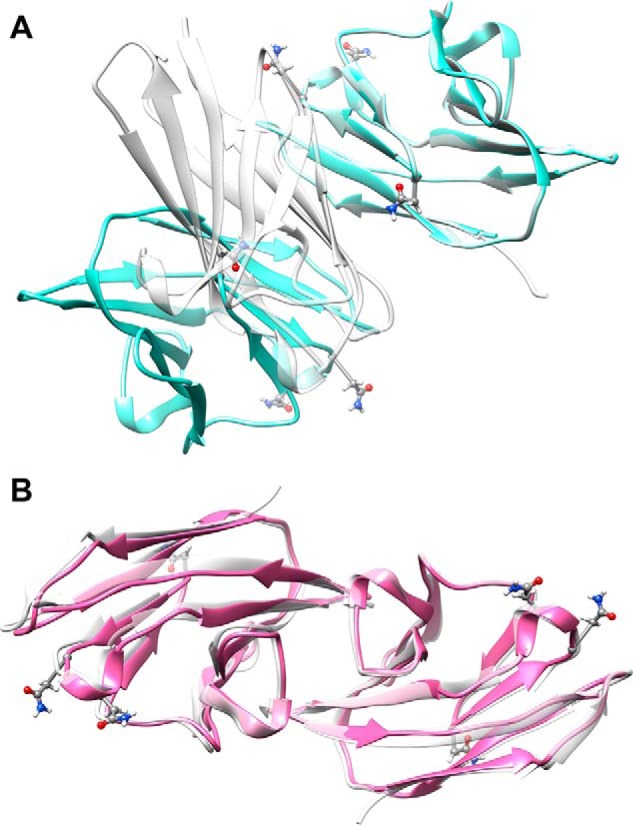
Non-glycosylated CEACAM1-IgV dimers constructed by rotation of monomers about the symmetry axis identified from RDC data. A, the rotated monomer has been translated to produce a model (shown in blue) with a best match to the dimer interface shown in the crystal structure 2GK2. B, the rotated monomer has been translated to produce a model (shown in red) with a best match to the dimer interface dimer shown in the crystal structure 4QXW. In each case the crystal structure dimers are shown in gray. One monomer of the model has been matched to a monomer in the crystal structure to depict the deviation in the position of the second monomer. The backbone root mean square deviation between the non-superimposed monomer is 19.7 Å in A and 1.0 Å in B. The glycosylation sites are represented in ball-and-stick mode for the asparagine residues.
