FIGURE 1.
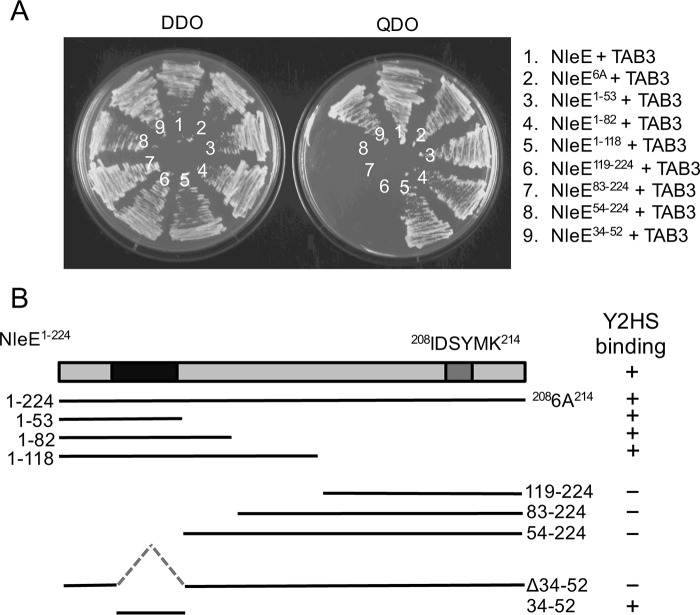
Mapping of NleE-TAB3 interaction domains. A, growth of S. cerevisiae AH109 on medium lacking histidine, adenine, leucine, and tryptophan (QDO) to select for protein-protein interactions (right panel) or medium lacking leucine and tryptophan (DDO) to select for plasmid maintenance only (left panel). Yeast are co-expressing NleE and TAB3 (1), NleE6A and TAB3 (2), NleE1–53 and TAB3 (3), NleE1–82 and TAB3 (4), NleE1–118 and TAB3 (5), NleE119–224 and TAB3 (6), NleE83–224 and TAB3 (7), NleE54–224 and TAB3 (8), and NleE34–52 and TAB3 (9). B, schematic representation of NleE and various deletion profiles with TAB3 binding capacity indicated in the right panel. Y2HS, yeast two-hybrid system.