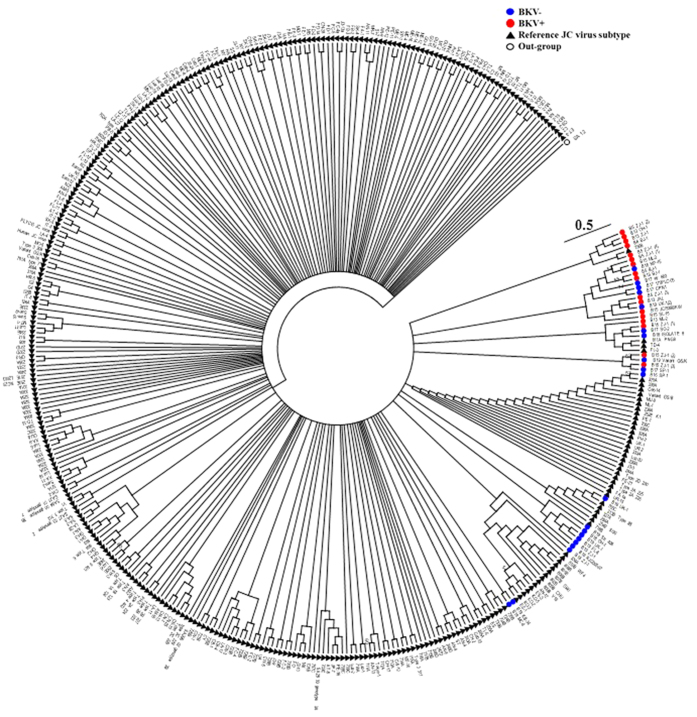
Figure 5. Phylogenetic analysis JC virus.
Phylogenetic tree based on the reference genomes of 333 JCV strains. The genomes from the BKV+ (red) and BKV− (blue) subjects are intercalated into the circular dendograms. The evolutionary history of the virus strains was inferred by the maximum likelihood method based on the Tamura-Nei model. The bootstrap consensus trees were inferred from 100 replicates that represent the evolutionary history of the virus taxa. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. SA12 (simian agent 12, a baboon polyoma virus) was used as an out group. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Samples analyzed in the study are labelled as - sample name/isolate name.