Figure 1.
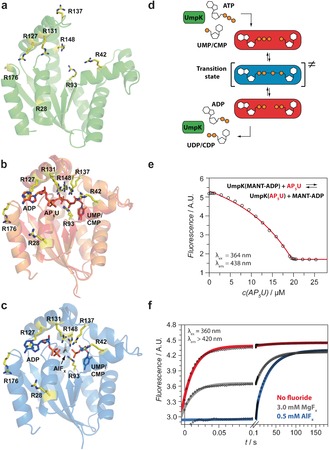
a) Nucleotide‐free UmpK (H. sapiens, PDB: 1TEV10). b) Overlay of UmpK in complex with the bi‐substrate inhibitor AP5U, (red, D. discoideum, PDB: 1UKE11), and in complex with ADP and CMP (orange, D. discoideum, PDB: 2UKD7). c) UmpK in complex with the TSA ADP:AlFx:CMP (D. discoideum, PDB: 3UKD7). In (a–c) arginine side‐chains are shown in yellow (numbering refers to UmpK from D. discoideum). d) Schematic representation of the reaction catalyzed by UmpK and the states studied here: nucleotide‐free (green), substrate/product‐bound (red), and the transition state (blue). e) AP5U binding to UmpK monitored by equilibrium displacement of fluorescent MANT‐ADP. Binding of AP5U (K D≤1.0 nm) generates a stable substrate/product‐like state. f) Nucleotide exchange kinetics measured by stopped flow. UmpK incubated with UMP and ADP in the presence of 0.5 mm AlFx (blue), 3 mm MgFx (grey) or in the absence of any fluoride (red) was mixed rapidly with the competing ligand MANT‐AP5A‐MANT. Notably, the dissociation of ADP and UMP is 2000‐fold slower in the presence of AlFx (k slow≈0.03 s−1) than in the absence of metal fluoride (k fast≈60 s−1) and the complex with AlFx is characterized exclusively by slow nucleotide exchange.
