Figure 3.
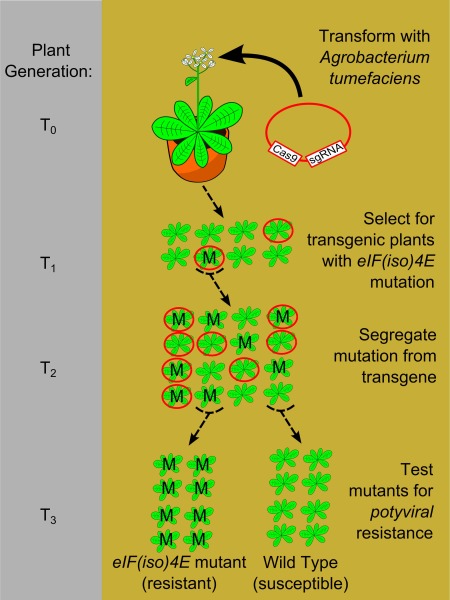
Schematic diagram of project workflow. Arabidopsis Col‐0 plants were transformed by floral dipping with the Cas9/sgRNA recombinant binary vector containing a BASTA resistance gene [pDe‐Cas9‐sgAteIF(iso)4E]. After self‐pollination, the seeds were collected and germinated in soil. Plants carrying the transgenic construct (red circle) were selected in the T1 generation by spraying with BASTA. The transgenic plants were then tested for eIF(iso)4E mutation (M) to identify plants with an active CRISPR/Cas9 nuclease using a T7 assay. One transgenic T1 plant with clear signs of eIF(iso)4E editing was used to produce the T2 generation. Polymerase chain reaction (PCR) was used to identify T2 generation plants which had lost the transgene by Mendelian segregation. The non‐transgenic T2 plants were then screened for eIF(iso)4E mutations by Sanger sequencing. (Please note: the mutations depicted as ‘M’ in the diagram are not identical, as the mutations in the T1 generation occurred in somatic cells, and so were not heritable, and different mutations were recovered in the T2 generation because of independent editing events in the germline of T1. For simplicity, ‘M’ was used to depict all CRISPR/Cas9‐induced mutations.) Non‐transgenic T2 plants, which were homozygous for either the mutated or wild‐type eIF(iso)4e alleles, were used to produce T3 populations, which were then tested for viral resistance.
