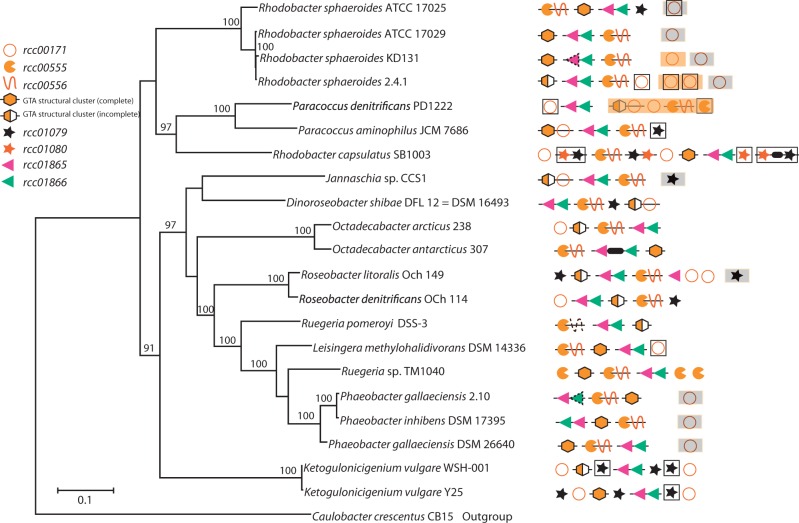
Fig. 6.
Conservation of the GTA loci across Rhodobacterales genomes. The presence and relative locations of RcGTA gene homologs are mapped onto the reference phylogenetic tree of the 21 analyzed Rhodobacterales genomes. The GTA “genome” loci are arranged as they appear in a genome relative to the annotated origin of replication. Adjacent genes are connected by lines. Genes that are found on a different chromosome or a plasmid are shown with orange and gray backgrounds, respectively. Regions outlined by black lines are located within or close to (<5 ORFs) predicted prophages. Symbols with a dotted contour indicate regions where the complete ORF is not present, but significant sequence similarity was nevertheless detected. A thick black line between genes represents the presence of an intervening ORF. An “incomplete” GTA structural cluster lacks at least one of the rcc01682–rcc01698 homologs. The reference phylogenetic tree was reconstructed by maximum likelihood from a concatenated alignment of 32 core genes. Caulobacter crescentus CB15 was used as an outgroup. Only bootstrap support values >70% are shown.