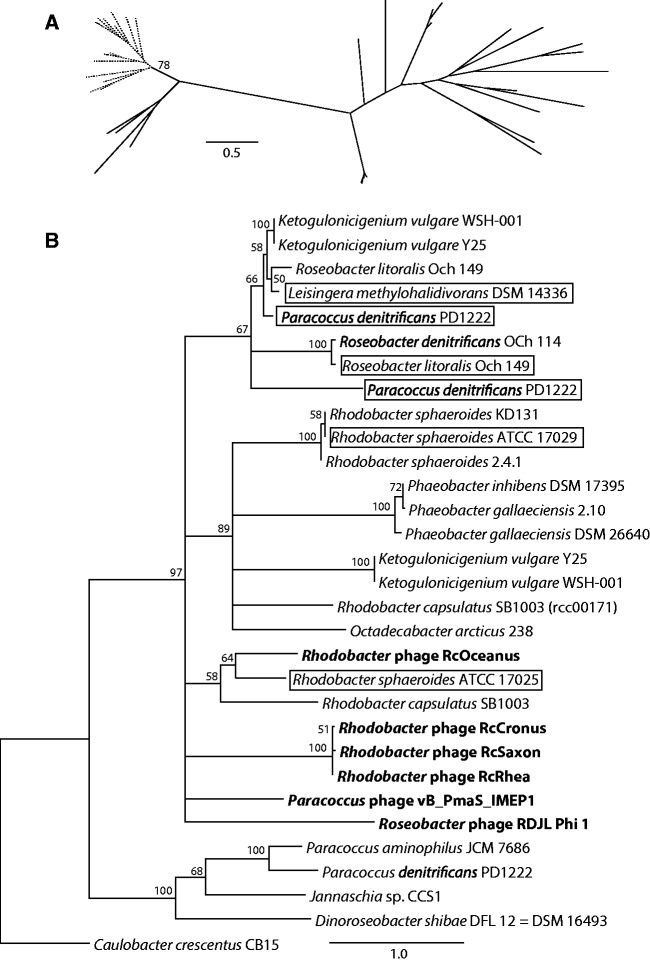
Fig. 7.
Examples of evolutionary relationships of phage and Rhodobacterales GTA gene homologs. (A) The phylogeny of a gene from within the structural cluster, rcc01696 (orfg13). All Rhodobacterales (branches in dotted lines) group separately from the bona fide phages (branches in solid lines). Bootstrap support is only shown for the bipartition separating the Rhodobacterales and phage homologs. (B) The phylogeny of a gene outside of the structural cluster, rcc00171. Phage homologs (taxa names in bold font) group within the Rhodobacterales, and the relationships among the Rhodobacterales homologs are incongruent with the reference tree (fig. 6). Outlined taxa likely represent homologs of phage origin, as they were found within or close to (<5 ORFs) a predicted prophage region. The tree was rooted using the Caulobacter crescentus CB15 sequence as an outgroup. Only bootstrap support values ≥50% are shown, and branches with <50% support were collapsed. Scale bars indicate the number of substitutions per site.