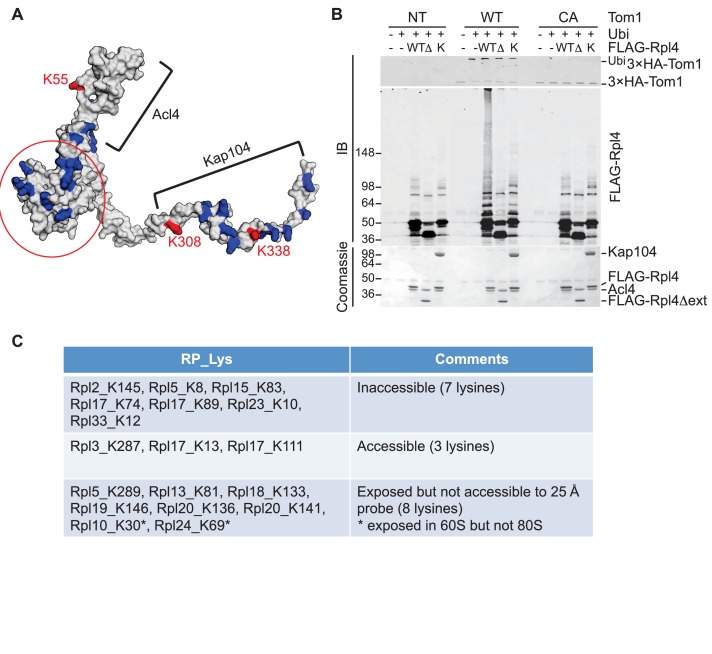
Figure 5. Tom1 acts through residues that are normally inaccessible in the structure of the mature ribosome.
(A) Structure of Rpl4 within the mature ribosome (PDB ID 4V88). Lysine residues are colored blue, with K55, K308, and K338 colored red. Areas involved in binding Acl4 and ctKap104 are indicated. The exact boundaries of the Kap104 binding site are not known. The globular central domain, which is fully exposed in the ternary Acl4–Rpl4–Kap104 complex but is not ubiquitinated, is circled in red. (B) Ubiquitination of Acl4–Rpl4 by 3xHATom1. Anti-HA immunoprecipitates from untagged (NT), 3xHATOM1 (WT), and 3xHAtom1CA (CA) cells were supplemented or not with E1/E2/ubiquitin/ATP (Ubi) and purified Acl4–FLAGRpl4, Acl4-FLAGRpl4∆ext and Acl4–FLAGRpl4–ctKap104 proteins. Samples were analyzed by SDS-PAGE and staining with Coomassie blue or immunoblotting with the indicated antibodies. WT, ∆, and K refer to Acl4–FLAGRpl4, Acl4-FLAGRpl4∆ext and Acl4–FLAGRpl4–ctKap104, respectively. See detailed methods in Material and methods. n = 3 biological replicates. (C) Tom1 preferentially targets lysines that are inaccessible in mature ribosomes. Lysine residues shown are those from large subunit ribosomal proteins in Figure 3E that are incorporated in the model for the structure of the yeast ribosome (pdb 4V88). The structure of a HECT domain–donor ubiquitin complex (pdb 4LCD) predicts that a gap of radius 25 Å must be present for Tom1 to access a lysine for ubiquitination. Two of the sites (Rpl10 K30 and Rpl24 K69) are accessible in the 60S large subunit but become inaccessible upon formation of the 80S ribosome.