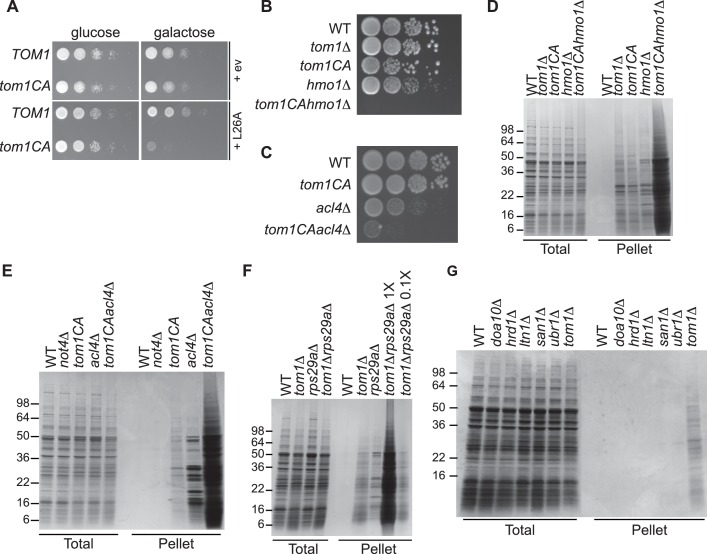
Figure 6. Defective ribosome assembly homeostasis and proteostatic collapse in tom1 mutant cells.
(A–C) Hypersensitivity of tom1CA cells to imbalances in ribosome components. (A) Cells of the indicated genotypes were spotted in serial 10-fold dilutions on glucose or galactose medium and incubated at 30°C for 2 days. ev refers to empty vector. n = 2 biological replicates. (B, C) As in (A) except that cells of the indicated genotypes were spotted on YPD. n = 2 biological replicates. (D–G) Massive accumulation of insoluble proteins in tom1 mutant cells. Cells of the indicated genotypes were lysed and fractionated into detergent-soluble and insoluble fractions, which were separated by SDS-PAGE and stained with Coomassie Blue. The pellet fraction is overloaded 20-fold compared to the total and supernatant fractions. n = 2 biological replicates.