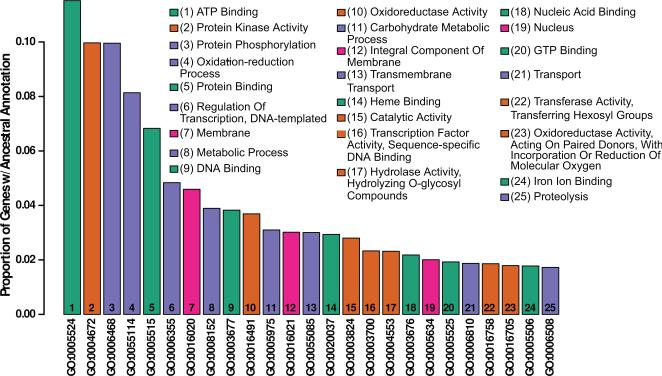
Figure 1.
D. capensis genome annotation. The 25 most common functional annotations either directly associated or ancestral to directly associated annotations on MAKER-identified genes in D. capensis. The fraction of identified genes associated directly or indirectly with each annotation code is shown on the vertical axis, with annotations shown in descending order of frequency. The description field of each GO code is shown (inset) by rank order. Hierarchical clustering of intercode distances within the ontology network was used to group the 25 most common functional annotations into four classes (denoted by color): ligand binding (aqua); membrane or nuclear location (magenta); and two groups of enzymatic functions (orange and violet).