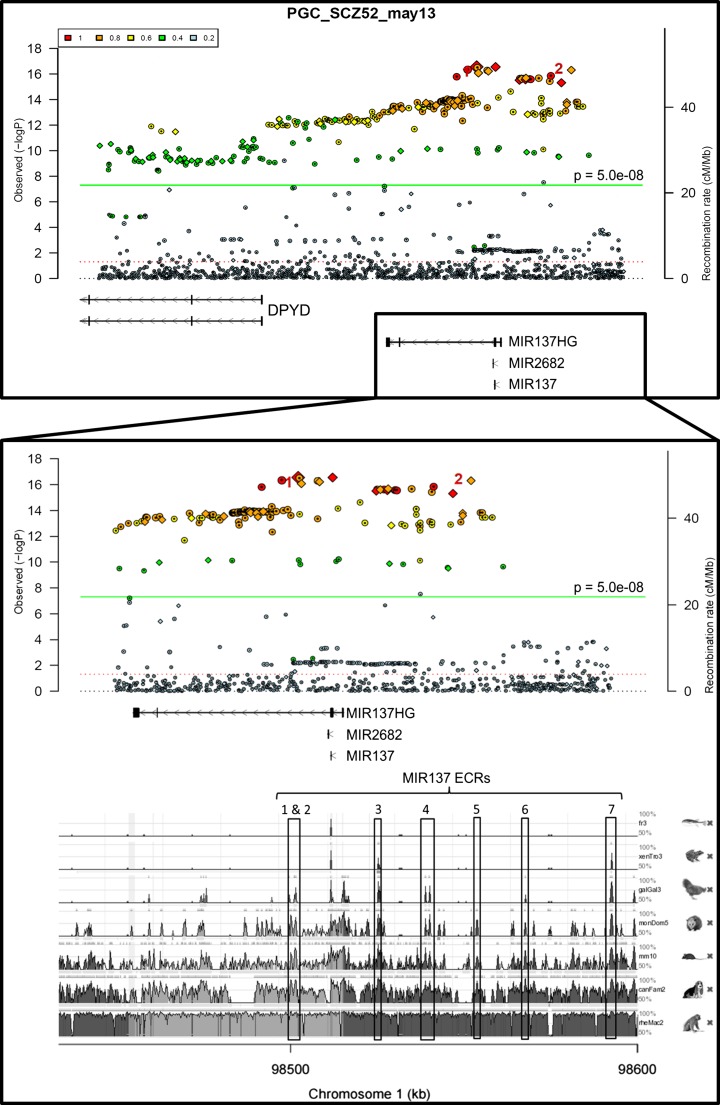
Fig. 1.
Schizophrenia genome-wide associated SNPs and evolutionary conserved regions (ECRs) at the MIR137 locus. Visualisation of the Psychiatric Genomics Consortium’s May 2013 schizophrenia GWAS data across the full schizophrenia-associated locus (chr1:98298371-98595000, GRCh37/hg19), using the Broad Institute’s Ricopili tool. The boxed region including MIR137 and upstream regions (chr1:98448000-98595000; GRCh37/hg19) displays the highest signal at this locus in terms of association for schizophrenia, suggesting that MIR137 and potential upstream regulatory regions are likely to be the causal association at this locus. The boxed region is expanded below, and schizophrenia GWAS SNP data overlaid onto species conservation data from the ECR browser. Statistically significant schizophrenia GWAS SNPs are represented by coloured circles or diamonds above the green line (p = 5.0e-08). Red numbers 1 and 2 highlight the schizophrenia GWAS SNPs rs1625579 and rs1198588, respectively. ECRs selected for study are boxed and numbered. Conserved peaks between boxes 2 and 3 represent the microRNA and its proximal promoter, which were not included in this analysis, but highlight conservation as an indicator of functionality