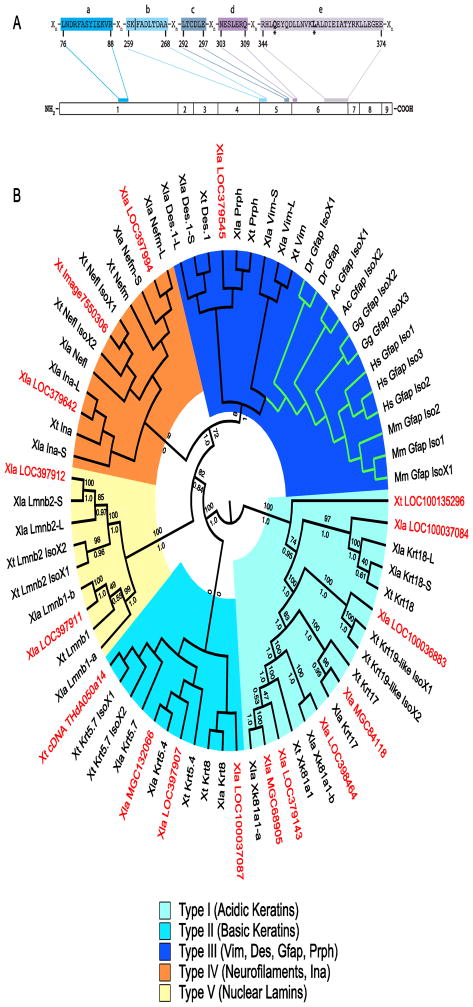
Figure 2. Sequences most similar to GFAP in Xenopus.
A GFAP consensus used to distinguish candidate GFAP sequences from other intermediate filament proteins. Conserved regions (labelled a–e) are present in all GFAP orthologs identified and separated by regions of varying length (Xn). The gfap exon in which each region is coded is shown. The numbering system below the consensus is for human GFAP Isoform 1 (NP_002046). All residues shown are invariant in the GFAP orthologs aligned (Figure S1). The residues (*) at positions 347 (Q) and 357 (L) are unique to GFAP. Consensus region b is coded for in two exons (4 and 5). Vertical line (|) demarcates sequence coding for exons 4 and 5. B Cladogram of GFAP orthologs, Xenopus IFPs and untitled Xenopus proteins with similarity to GFAP. Midpoint rooted Polar Tree based on MAFFT alignment of GFAP orthologs, selected known X. laevis and X. tropicalis IFPs and the 17 untitled X. laevis and X. tropicalis sequences showing greatest similarity to GFAP (Katoh and Standley, 2013). Untitled X. laevis (Xla) and X. tropicalis (Xt) sequences are shown in red. Green branches show location of the GFAP clade. Groupings of IFP types are shown. Maximum likelihood bootstrap (above) and Bayesian inference posterior probabilities (below) for each branch are included (Felsenstein, 1981; Huelsenbeck et al., 2001). Accession numbers for each sequence in the tree can be found in Supplementary Table S2.