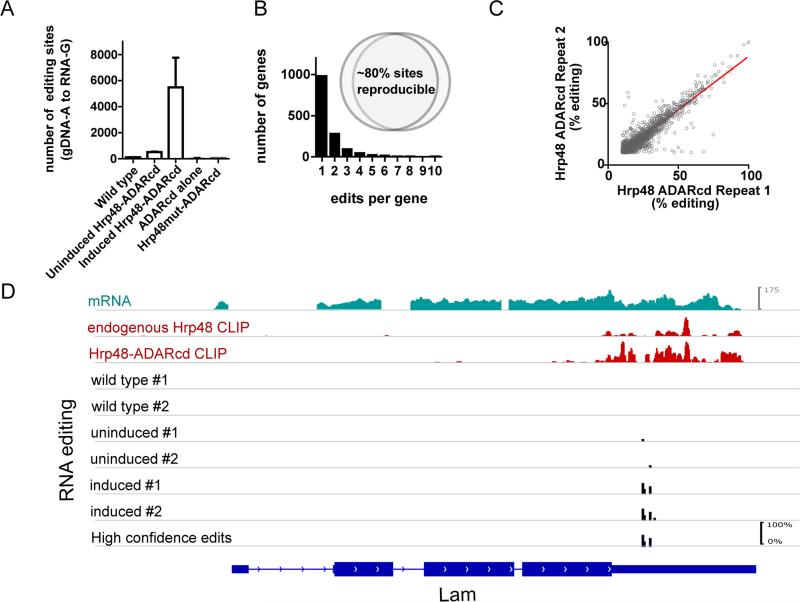
Figure 2. The TRIBE fusion protein reproducibly edits certain sites.
(A) An increase in A to G editing events is observed upon induction of the fusion protein in S2 cells. No increase in editing sites are observed when an ADAR catalytic domain alone is expressed, or when Hrp48mut-ADARcd (Hrp48 with mutated RNA-binding domains). The same genes and the same sites are reproducibly edited across biological replicates at similar efficiencies (B inset, C). A frequency histogram of number of edits per target genes show that most genes have only one editing site but the TRIBE protein has strong specificity for certain sites (B), and that those sites are edited to a similar degree between biological repeats (R2 = 0.859). D) Endogenous and fusion protein have similar binding patterns and TRIBE editing reflects the pattern of the CLIP signal. An example gene, Lam, showing mRNA expression and CLIP signal (top three panels), and editing tracks for wild type cells, stable cells lines (Hrp48-TRIBE) without and with induction of expression of the fusion protein. Editing events are indicated by black bars, the height of the bar indicates the percentage editing at that site.