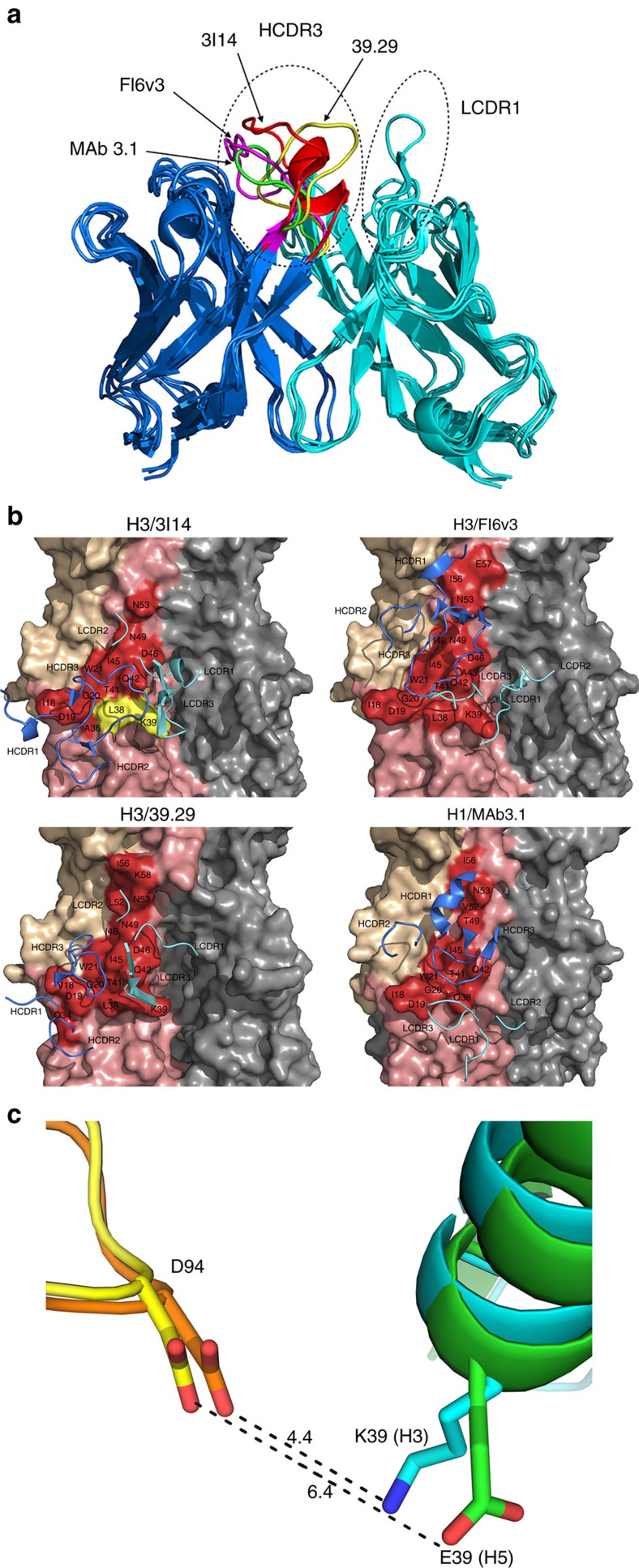
Figure 6. Modelling of 3I14 and docking with H3/H5.
(a) The superimposition of 3I14 model with FI6v3, 39.29 and MAb 3.1. The bnAbs are displayed in ribbon representations. The heavy chain is in blue and the light chain is in cyan. The HCDR3s and LCDR1s are indicated by the hatched ovals. The residues in HCDR3 of 3I14, FI6v3, 39.29 and MAb 3.1 are coloured in red, magenta, yellow and green, respectively. (b) The complex structures of IGHV3-30-derived bnAbs with HAs. The epitope residues on the HAs are displayed in surface representation and the CDR loops of bnAbs are shown are shown as ribbons. HA1 of HA monomer is in wheat, HA2 is in salmon and epitope residues are labelled as red. The key residues L38 and K39 are coloured in yellow. Heavy chain CDRs of bnAbs are in blue and light-chain CDRs are in cyan. 3I14 was homology modelled using the antibody-modelling module in BioLuminate and the model was superimposed to H3/39.29 complex structure before docking with RosettaDock. Other three IGHV3-30 bnAbs, FI6v3, 39.29 and MAb 3.1 were downloaded from Protein Data Bank. (c) The interaction of D94 in 3I14 with H3/H5. H3 is shown in cyan with K39 shown as stick; H5 is shown in green with E39 shown as stick; 3I14 is shown in orange in H3/3I14 model and in yellow in H5/3I14 model with D94 shown as stick.