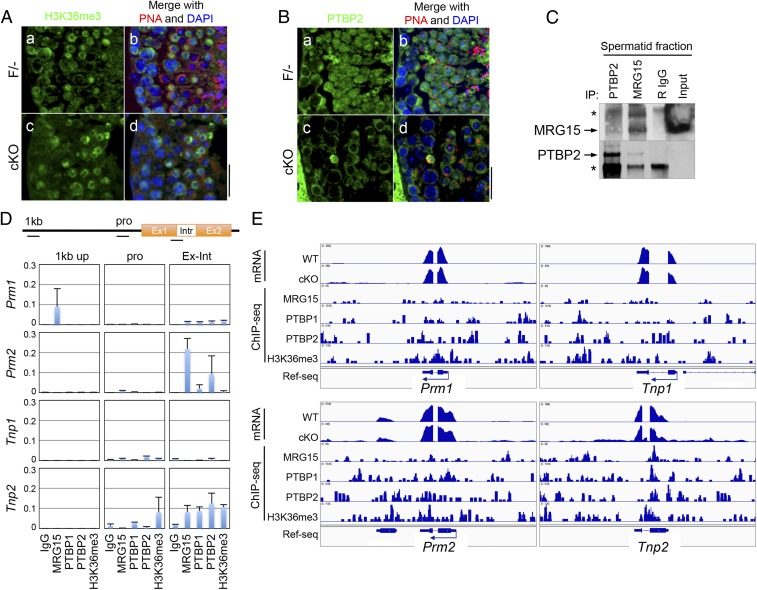
Fig. 5.
MRG15 and PTBP2 cooperatively regulate splicing of Tnp2 gene. (A) Accumulation of H3K36me3 in the testes. (a and c) H3K36me3 (green) and (b and d) peanut agglutinin (PNA; red) were visualized in the (a and b) F/− and (c and d) cKO testes. (B) Immunolocalization of PTBP2. (a and c) PTBP2 (green) and (b and d) PNA (red) were stained in the (a and b) F/− and (c and d) cKO testes. (Scale bar: 50 µm.) (C) Coimmunoprecipitation of MRG15 and PTBP2 in WT spermatid fraction. Arrows indicate the bands of each protein. IP, immunoprecipitation; R, rabbit; *, IgG. (D) ChIP analyses of the protamine and transition protein genes. Quantitative PCR using primers located on 1-kb upstream, promoter, and exon–intron junction of Prm1, Prm2, Tnp1, and Tnp2 (each location is shown in Upper) was performed in ChIPs with indicated antibodies. Results are shown as relative amplification against input genome. (E) Colocalization of retained intron and MRG15 complexes. Genome browser views show the alignment of the read sequences of mRNA and ChIP with MRG15, PTBP1, PTBP2, and H3K36me3 on the mouse genomic regions of Prm1, Prm2, Tnp1, and Tnp2 of WT spermatids. Ref-seq, reference sequence.