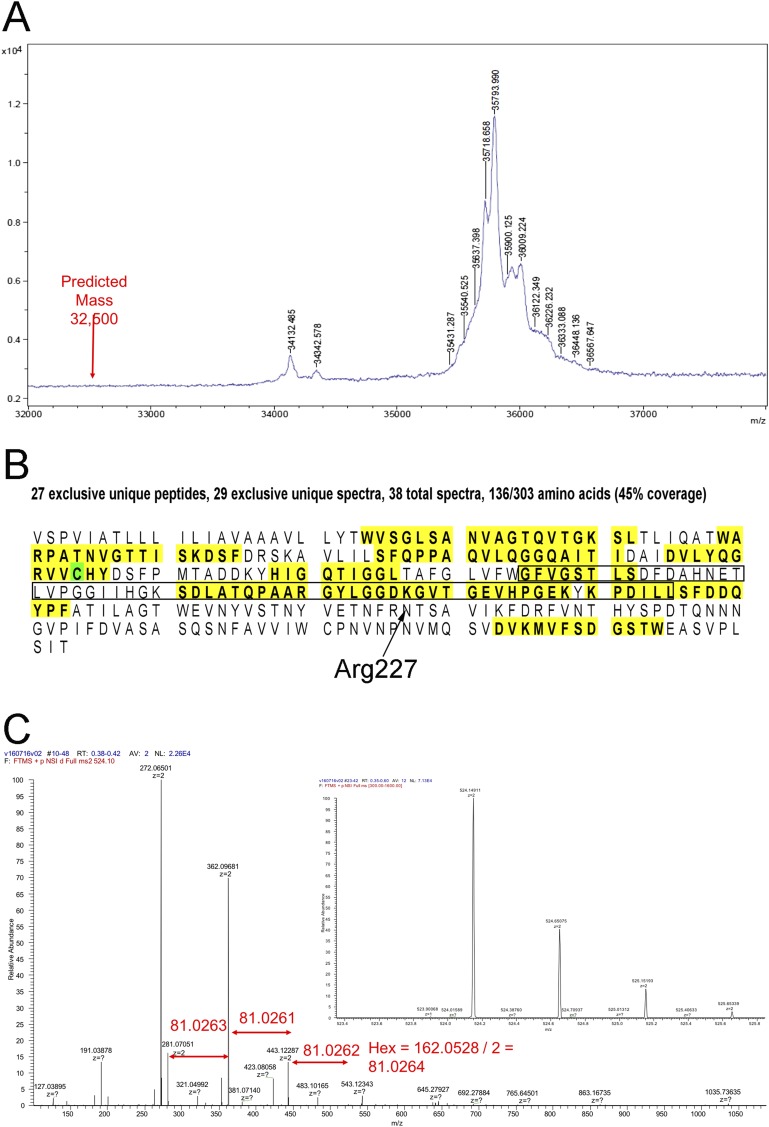
Fig. S5.
MS of Iho670. (A) MALDI-TOF mass analysis showing that the actual mass is from 1 to 4 kDa higher than that predicted from the protein sequence. The single largest species (35,794 Da) has a mass that is ∼3.3 kDa greater than expected. (B) Digestion of the protein (yellow) shows the unmodified peptides that were identified. Cys104 (green) was determined to be modified with a carbamidomethyl group (57 Da). The region that we were unable to trace (residues 135–195) in building an atomic model is shown within a box. (C) Fragmentation analysis of several ions showed that mass loss involved the removal of hexose, establishing that glycosylation was present.