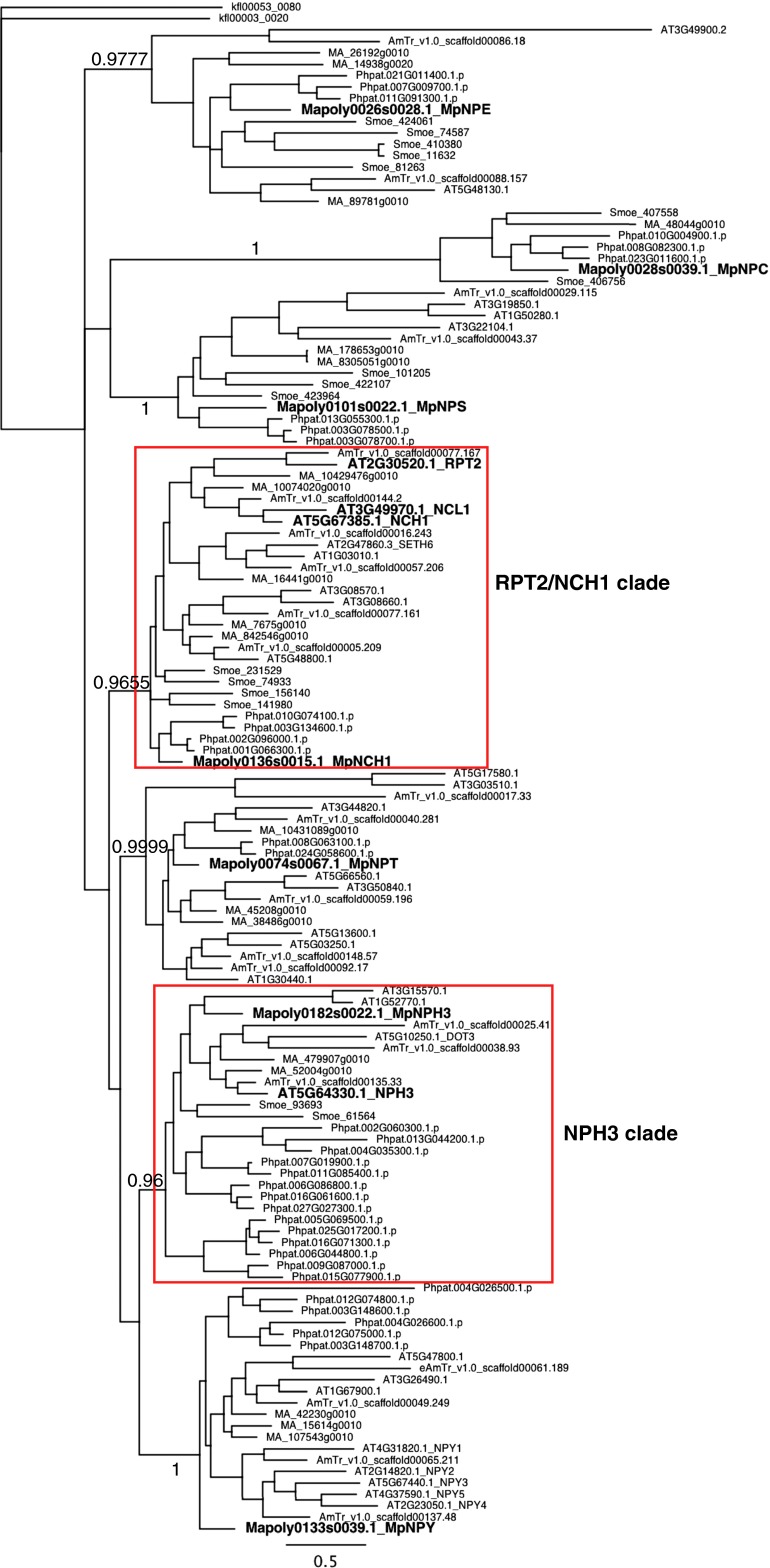
Fig. S2.
The Bayesian phylogenetic tree of NRL proteins. The parameters used were as follows: preset amino acid model program, mixed (LG, blosum, and wag); length of set rates, inverse-gamma; number of gamma categories, 4; Markov-chain Monte Carlo start tree, random; number of generation, 1,000,000; number of runs, 2; number of chains, 4; heated chain temperature, 0.2; sample frequency, 200; and burn-in fraction, 0.25. klfl00053_0080 were used as the outgroup. The numbers at the nodes indicate posterior probabilities. Bars indicate substitution values per site. Both RPT2/NCH1 and NPH3 clades are indicated (red box). Seven Marchantia and four Arabidopsis NRL proteins (NCH1, RPT2, NCL1, and NPH3) are bold-faced. AT, Arabidopsis thaliana; AmTr, Amborella trichopoda; MA, Picea abies; Smoe, Selaginella moellendorfii; Phpat, Physcomitrella patens; Mapoly, Marchantia polymorpha; and kfl, Klebsormidium flaccidum.