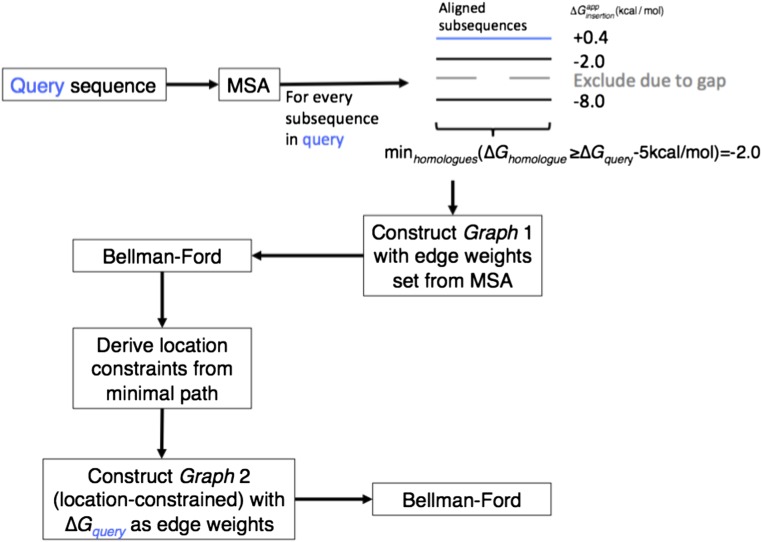
Fig. S2.
TopGraphMSA algorithm flowchart. (A) Nonredundant MSA is generated for a query sequence using BLAST, CD-HIT clustering, and MUSCLE alignment. For every possible segment is calculated. Values different from the query’s by more than 5 kcal/mol are discarded. The minimal value found is used as the weight for all edges leading to the segment’s node. The Bellman–Ford algorithm is applied to find the minimum energy topology. Using the minimal topology, constraints are derived for every chosen segment. A graph is rebuilt such that every segment found in the MSA is a constraint, and weights are assigned as the of the corresponding query sequence. The Bellman–Ford algorithm (28) is used again to find the minimal energy topology.