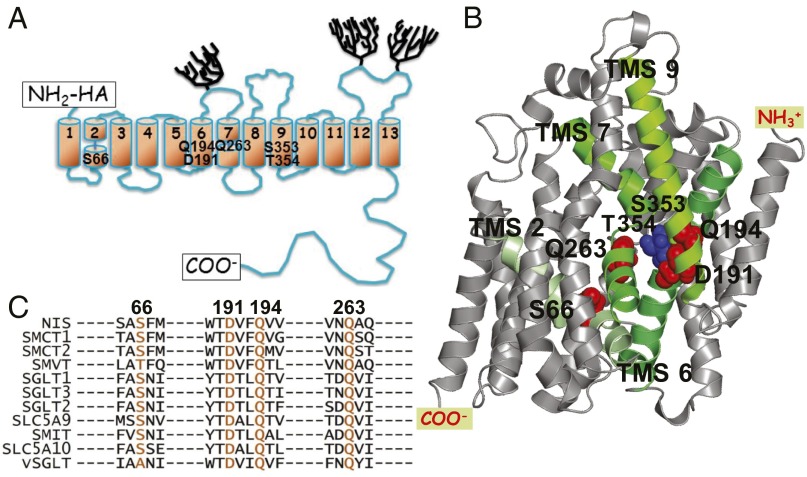
Fig. 1.
NIS models and SLC5 family alignment. (A) NIS secondary structure model. Numbered cylinders represent the transmembrane segments (TMSs); trees, the carbohydrates; and rectangles, the carboxy terminus and the HA tag at the amino terminus. The positions of the residues determined to be significant for Na+ coordination at the Na2 site are identified by residue name. (B) NIS 3D homology model (5). Green TMSs are those containing the residues that interact with Na+ at the Na2 site. (C) The residues named in A are highly conserved in the SLC5 family. The residues in the other members of the family that correspond to the NIS residues of interest are shown in orange. Alignment was generated using ClustalW2 (www.ebi.ac.uk/Tools/msa/clustalw2/).