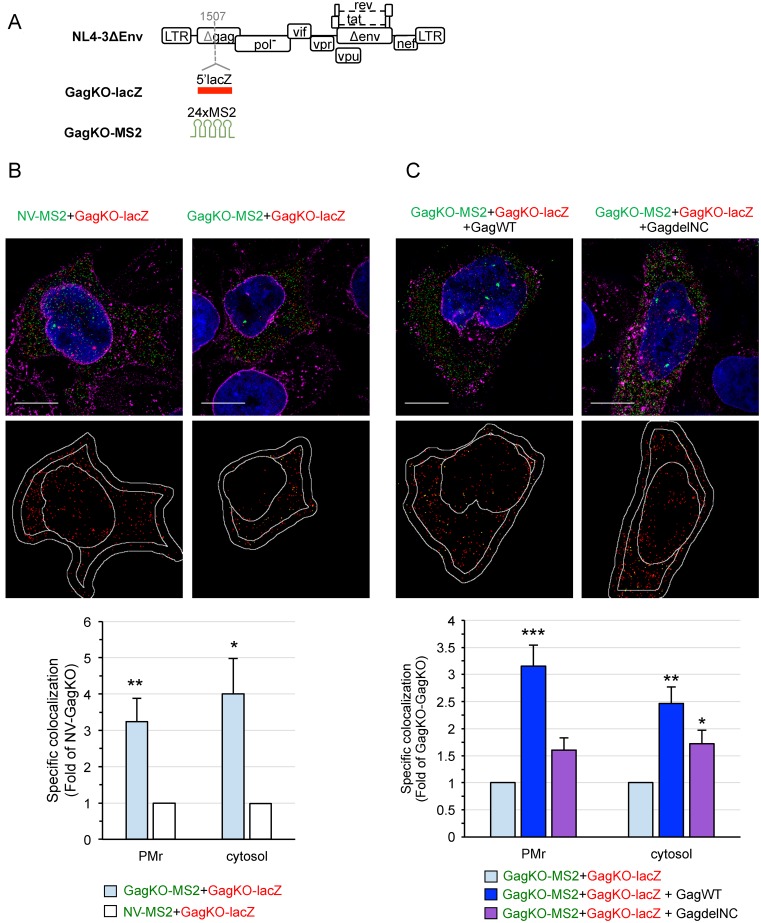
Figure 6.
3D-SIM analysis of the role of Gag in HIV-1 gRNA colocalization. (A) Schematic representation of the constructs used in this study. To generate GagKO mutants, lacZ or MS2 tags were inserted within the capsid domain of the gag gene (position indicated by a dashed gray line). The insertion prevents the synthesis of Gag and Gag-Pol polyproteins but maintains the expression of all the regulatory and accessory proteins. Representative 3D-SIM images of cells co-transfected with GagKO-lacZ (red) and GagKO-MS2 (green) in the absence (B) or the presence (C) of Gag (GagWT or GagdelNC). The lower images show the centers of red spots (red) and those that colocalized with a green spot in yellow. Scale bars are 10μm. The graphs show the specific colocalization normalized to the control (GagKO-lacZ+NV-MS2 in B) or to GagKO-MS2+GagKO-lacZ (C). Data are the mean ± SEM of 30 cells shown for each condition. The significance of differences with control (NV-GagKO in B) or with GagKO-GagKO (C) was assessed using an unpaired Student's t-test (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001). No significant difference was found between the trans-complementation with GagWT or GagdelNC in the cytosol.