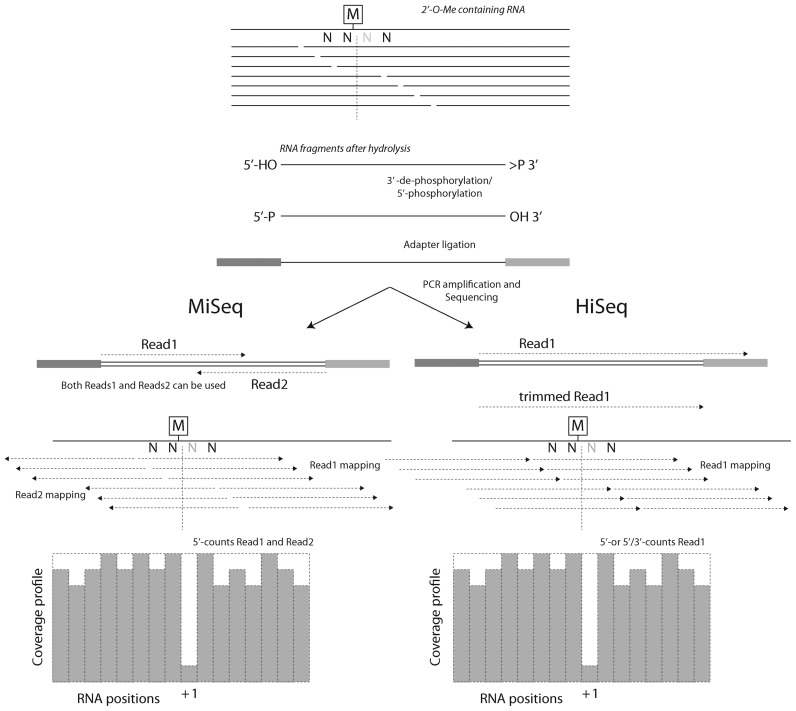
Figure 1.
General overview of the RiboMethSeq protocol. RNA containing 2′-O-Me residues is randomly fragmented, 5′- and 3′-ends are repaired and adapters are ligated to both extremities. After amplification and barcoding, amplicons are subjected to Illumina sequencing. MiSeq sequencing (left) generally provides paired-end reads, while HiSeq sequencing is generally performed in a single-read mode (right). Orientations and mapping of reads are indicated. The 2′-O-Me residues protect the 3′-adjacent phosphodiester bond from cleavage, generating a typical gap in 5′-/3′-ends coverage profile.