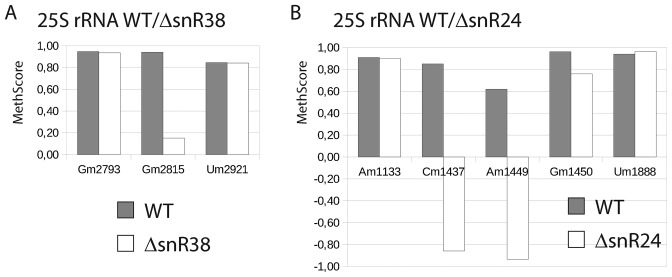
Figure 5.
Validation of the RiboMethSeq approach using yeast deleted strains deficient in 2′-O-methylation residue at certain positions in 25S rRNA. Two yeast strains with deletions of snR24 and snR38, together with the corresponding WT strain, were subjected to RiboMethSeq. Variations of MethScore values are observed upon deletion of snR38, responsible for modification of position Gm2815 (left) and snR24, implicated for modification of positions Cm1437, Am1449 and Cm1450 (right). Two neighboring positions around of the affected modification site(s) are also shown for comparison. In gray, values obtained for WT yeast strain, in white, values for corresponding snoRNA-deleted mutant.