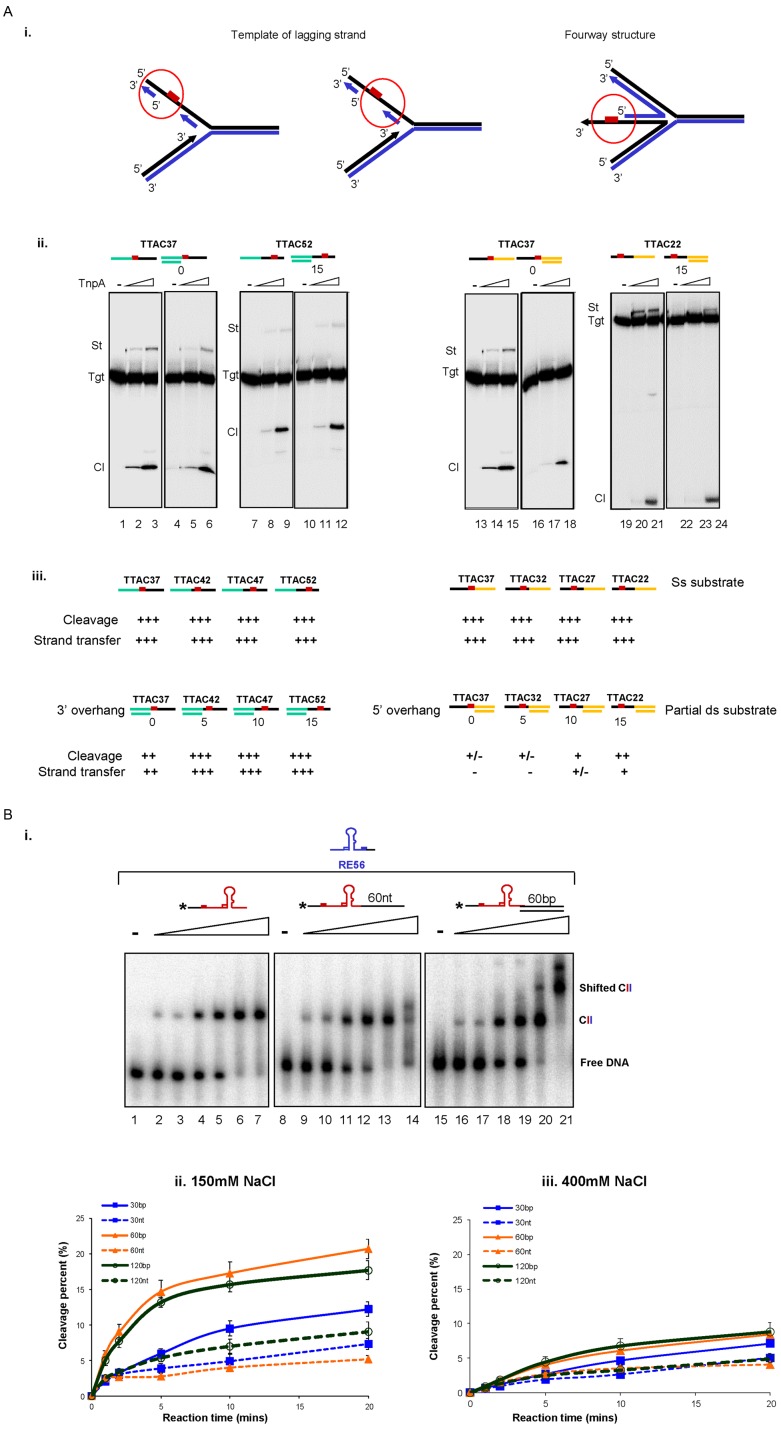
Figure 5.
Effect of dsDNA neighbouring to ssDNA on integration and excision. (A) The dsDNA neighbouring to target site affected the integration activity. (i) Cartoon representing different ds-ss DNA regions at the replication fork. Red box represents the target TTAC. (ii) Integration activity on 3′ overhang (lanes 4–6 and 10–12) and 5′ overhang substrates (lanes 16–18 and 22–24) compared to ss substrates (lanes 1–3, 7–9 and 13–15, 19–21) respectively. (iii) Summary of TnpAIS608 relative cleavage and integration activities on ss and partial ds targets. (B) The presence of dsDNA adjacent to ss ends increases the rate of TnpAIS608-catalysed cleavage. (i) Binding of TnpAIS608 to ss0 (lanes 1–7), ss60 (lanes 8–14) and ds60 (lanes 15–21) substrates by EMSA. Increasing TnpAIS608-His6 concentrations (60, 150, 300, 450, 600 and 1000 nM) are indicated by the triangle. Complexes were separated in native 5% polyacrylamide gels. (ii) Cleavage kinetics in 150 mM NaCl with standard reaction conditions. (iii) Cleavage kinetics in 400 mM NaCl.