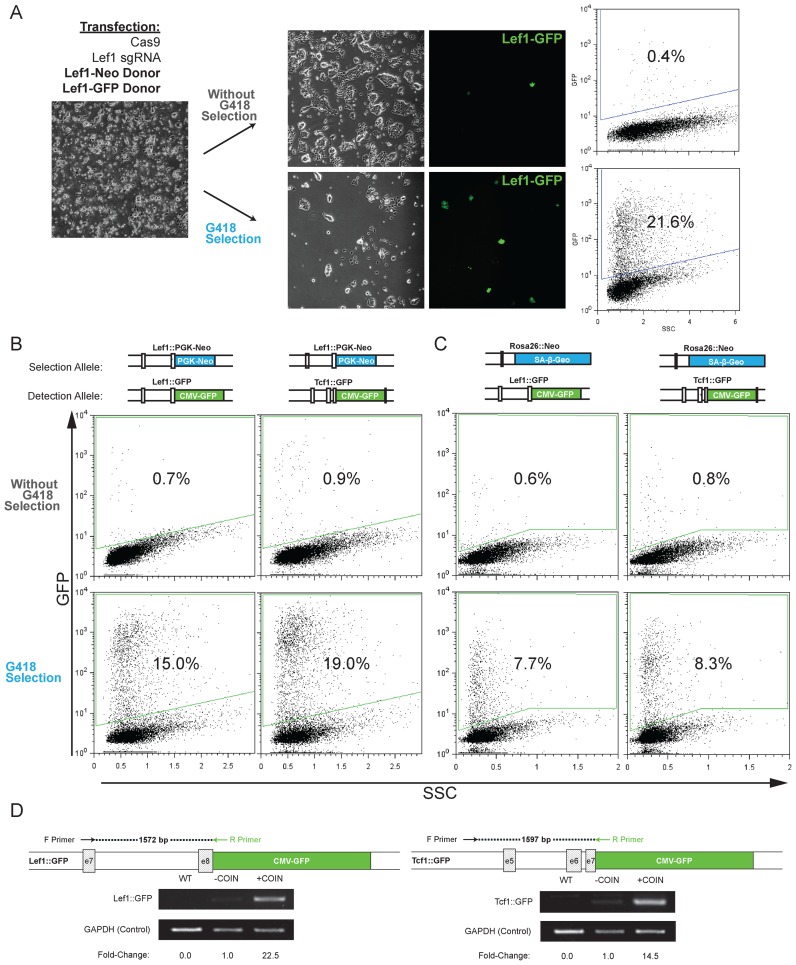
Figure 3.
High frequency of coincidental insertion of distinct donor DNA targeting the same gene and independent genes. (A) Experimental approach showing transfection of donor DNA for selection (Lef1-PGK-Neo) and detection (Lef1-CMV-GFP), splitting cells into G418-containing media or non-selective media, and detection of Lef1::GFP frequency by microscopy and flow cytometry to determine percentage of GFP+ cells. Each donor DNA had 208/202bp homology arms. The insertion percentage is determined by flow cytometry by measuring the total GFP+ cells after gating on a transfected, GFP-negative control population. (B and C) Similar experimental set as shown in (A); however, sgRNA and donor DNA target insertion at distinct genes (Lef1, Tcf1, Rosa26). The combination of selection and detection donor DNA are noted at the top of each column. The insertion percentage is measured by counting total GFP+ cells by flow cytometry after gating for a transfected, GFP-negative control population. Representative microscopy images are shown in Supplementary Figure S3A. (D) Semi-quantitative PCR analysis of COIN for Lef1 and Tcf1 loci was performed to confirm that coincidental insertion increases on-target integrations. For each locus, two primers were designed; an external primer specific to the gene locus outside of the homology arms and an internal primer specific to the insert. Densitometry of the PCR products is used to compare the relative frequency of integration in a population of cells with and without COIN.