Abstract
The penetration of antibiotics in necrotic tuberculosis lesions is heterogeneous and drug-specific, but the factors underlying such differential partitioning are unknown. We hypothesized that drug binding to macromolecules in necrotic foci (or caseum) prevents passive drug diffusion through avascular caseum, a critical site of infection. Using a caseum binding assay and MALDI mass spectrometry imaging of tuberculosis drugs, we showed that binding to caseum inversely correlates with passive diffusion into the necrotic core. We developed a high-throughput assay relying on rapid equilibrium dialysis and a caseum surrogate designed to mimic the composition of native caseum. A set of 279 compounds was profiled in this assay to generate a large data set and explore the physicochemical drivers of free diffusion into caseum. Principle component analysis and modeling of the data set delivered an in silico signature predictive of caseum binding, combining 69 molecular descriptors. Among the major positive drivers of binding were high lipophilicity and poor solubility. Determinants of molecular shape such as the number of rings, particularly aromatic rings, number of sp2 carbon counts, and volume-to-surface ratio negatively correlated with the free fraction, indicating that low-molecular-weight nonflat compounds are more likely to exhibit low caseum binding properties and diffuse effectively through caseum. To provide simple guidance in the property-based design of new compounds, a rule of thumb was derived whereby the sum of the hydrophobicity (clogP) and aromatic ring count is proportional to caseum binding. These tools can be used to ensure desirable lesion partitioning and guide the selection of optimal regimens against tuberculosis.
Keywords: Mycobacterium tuberculosis, granuloma, caseum, drug penetration, in vitro assay, principle component analysis
Graphical abstract
INTRODUCTION
The treatment of uncomplicated tuberculosis (TB) requires up to four drugs taken daily for 6 months, whereas most bacterial lung infections can be cured by a single antibiotic in 1 to 2 weeks. The complex TB pathology and its impact on bacterial physiology and drug penetration play critical roles in the relative inefficiency of anti-TB treatment.1–3
Because bacteria reside in both the cellular and necrotic regions of granulomas,4,5 effective penetration of drugs into the cellular layers and diffusion through the caseous center are likely essential to eradicating the infection. Poor drug penetration can lead to subinhibitory concentrations of TB drugs and periods of local monotherapy, which in turn increase the risk of the emergence of resistant mutants.2,6,7 In efficacy studies that focused on bacterial populations surviving drug treatment, the lesion compartments that failed to be sterilized at the end of therapy were mostly necrotic granulomas and caseous foci,4,8–10 where hypoxic conditions reduce the activity of many drugs. Clinically, cavitary TB, where large numbers of bacilli are found in the cavity caseum, is associated with inferior cure rates and poor prognoses.11,12 Thus, favorable distribution into the caseum of closed nodules and cavities is an important attribute of existing anti-TB agents and new drug candidates.
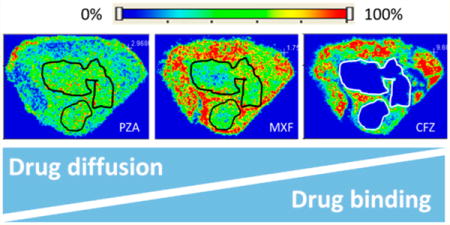
We have shown that patterns of drug penetration in rabbit and human necrotic lesions are strikingly drug-specific.2,13 The two-dimensional distribution of anti-TB agents was visualized by MALDI mass spectrometry imaging (MSI), revealing a wide range of diffusibility through the caseous foci of necrotic lesions.2,14 At one end of the spectrum, pyrazinamide diffused rapidly and extensively into caseum relative to the surrounding cellular area, and clofazimine showed very poor caseum penetration. The fluoroquinolones exhibited intermediate diffusion behavior in the necrotic region of TB lesions. Rifampicin, a drug with treatment-shortening and sterilizing properties, accumulated slowly in caseum and maintained therapeutic levels in this compartment throughout the dosing interval.2
Caseum has a “cheesy” appearance and is rich in cholesterol, cholesterol esters, triacylglycerols, and lactosylceramides.15 It originates from the necrosis and apoptosis of immune cells, mostly foamy macrophages in which lipid droplets have accumulated as a result of lipid metabolism dysregulation triggered by M. tuberculosis.15 This necrotic matrix is found at the core of closed granulomas and lining the walls of open cavities, which form when the central caseum liquefies and is evacuated through a lung airway. Granuloma formation and the life cycle of M. tuberculosis within lesions is extensively reviewed in refs 4–6. Most importantly, caseum is devoid of vascularization, leading to poor or no supply of fresh nutrients, oxygen, and drugs, but hosts a significant population of bacteria that have adapted to these hostile conditions. These bacteria are mostly extracellular, are slowly replicating or nonreplicating, and exhibit phenotypic drug tolerance as a result of hypometabolic adaptations.4,9,10,16
As a first step toward understanding the determinants of drug penetration into caseum, we developed and validated rapid equilibrium dialysis (RED) assays coupled to liquid chromatography and tandem mass spectrometry to measure the free fraction of drug-like molecules in caseum and caseum-like material. We report the profiling of a panel of small molecules using these assays. Principle component analysis (PCA) was applied to identify physicochemical and molecular predictors of experimental caseum binding. An in silico predictive model was developed to guide medicinal chemistry efforts in TB drug discovery as well as the rational design of combination therapies involving drugs that complement each other in their ability to penetrate caseous granulomas.
RESULTS
Association between Caseum Binding and Drug Partitioning in Vivo
Because necrotic material or caseum is entirely acellular and thus devoid of a vascular supply and active transport, only the free drug fraction can penetrate this matrix via passive diffusion. We hypothesized that factors influencing the fate of drug molecules at the cellular/necrotic interface would include intrinsic physicochemical properties and an extent of binding to caseum macromolecules. We first adapted the rapid equilibrium dialysis (RED) assay, commonly used to measure plasma protein binding, to determine drug binding to caseum. Building upon our previous results of MALDI mass spectrometry imaging (MSI) and conventional quantitation of drugs in TB lesions,2,17 we showed that in vivo drug diffusion through caseum is inversely correlated to binding to caseum macromolecules (Figure 1).
Figure 1.
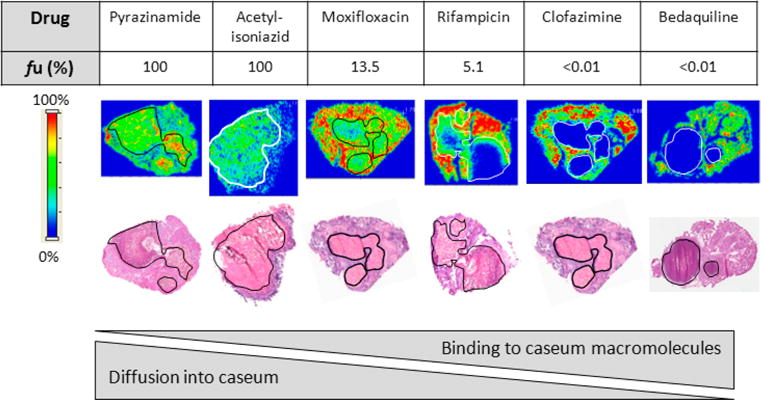
Drug penetration in vivo. Relationship between the fraction unbound (fu) and diffusion into caseum in vivo as determined by MALDI mass spectrometry imaging for six anti-TB drugs. Ion maps are of representative lung lesions, and the signal intensity is indicated by the scale bar to the left. Hematoxylin and eosin (H&E) staining of adjacent sections is shown below the ion maps. Black/white contour lines highlight the necrotic center of each lesion.
Next, we measured the plasma and caseum binding of 20 anti-TB drugs and metabolites in the RED assay, in which either naïve rabbit plasma or caseum homogenate from infected rabbits was used. To expand the range of physicochemical properties, additional TB-inactive compounds were included in the panel. The undiluted fraction unbound (fu) of all compounds is listed in Table 1. (See Table S1 for structures and physicochemical properties.) Caseum binding was highest for bedaquiline and clofazimine (fu < 0.01%), but there was no measurable binding of pyrazinamide and isoniazid (fu > 99.9%). Overall, we observed that compounds were more highly bound to caseum than to rabbit plasma and that plasma protein binding correlated poorly with caseum binding (Figure S1). We also observed significant differences in caseum binding between members of the same compound class. The fraction unbound of the five oxazolidinones tested (linezolid, posizolid, sutezolid, radezolid, and tedizolid) ranged from 5 to 30%. The three fluoroquinolones tested (moxifloxacin, levofloxacin, and gatifloxacin) had more comparable caseum binding (8% < fu < 16%). Discovery compounds with no antimycobacterial activity also exhibited a wide range of fractions unbound, from <1 to 56%.
Table 1.
Fraction Unbound (fu) of 36 TB Drugs and TB-Inactive Compoundsa
| rPlasma fu (%) | rCaseum fu (%) | surrogate fu (%) | |
|---|---|---|---|
| Ethambutol | 80.2 ± 5.5 | 35.2 ± 4.4 | 38.8 ± 5.6 |
| Isoniazid | >99.9 | >99.9 | >99.9 |
| Acetyl-isoniazid | 89.4 ± 4.7 | >99.9 | >99.9 |
| Pyrazinamide | >99.9 | >99.9 | 62.0 ± 3.5 |
| p-aminosalicylic acid | 43.9 ± 1.8 | 54.7 ± 1.4 | 51.1 ± 11.2 |
| Ethionamide | 29.6 ± 2.5b | 52.8 ± 2.4b | 27.8 ± 11.9 |
| Rifampicin | 6.05 ± 0.2 | 5.13 ± 0.2 | 7.3 ± 0.6 |
| Rifapentine | 0.59 ± 0.04 | 0.5 ± 0.1 | 1.1 ± 0.1 |
| Moxifloxacin | 64.5 ± 2.2 | 13.5 ± 3.7 | 16.8 ± 1.8 |
| Levofloxacin | 44.0 ± 6.0 | 8.34 ± 0.4 | 16.3 ± 3.1 |
| Gatifloxacin | 65.4 ± 5.1 | 16.3 ± 4.2 | 18.8 ± 2.9 |
| Linezolid | 56.7 ± 3.1 | 29.3 ± 3.6 | 27.9 ± 2.2 |
| Posizolid | 8.64 ± 0.9 | 10.7 ± 1.7 | 17.6 ± 4.5 |
| Sutezolid | 20.5 ± 0.2 | 30.1 ± 8.7 | 21.7 ± 1.7 |
| Radezolid | 3.05 ± 0.1 | 5.2 ± 0.8 | 7.6 ± 1.9 |
| Tedizolid | 30.7 ± 2.0 | 8.8 ± 1.8 | 13.7 ± 2.7 |
| Clofazimine | <0.01 | <0.01 | <0.01 |
| Bedaquiline | <0.01 | <0.01 | <0.01 |
| PA-824 | 11.1 ± 0.2 | 7.31 ± 2.2 | 3.6 ± 0.2 |
| OPC67683 | <0.01b | 0.04 ± 0.02b | 0.02 ± 0.002 |
| LLY001 | 44.7 ± 3.6 | 63.2 ± 7.3 | |
| LLY002 | 9.6 ± 0.8 | 15.3 ± 0.9 | |
| LLY003 | 31.8 ± 4.6 | 34.0 ± 5.1 | |
| LLY004 | 7.9 ± 0.4 | 22.8 ± 3.9 | |
| LLY005 | 26.8 ± 1.9 | 28.1 ± 6.2 | |
| LLY006 | 56.4 ± 6.6 | 79.05 ± 4.4 | |
| LLY007 | 4.1 ± 0.5 | 10 ± 2.6 | |
| LLY008 | 1.9 ± 0.1 | 1.78 ± 0.5 | |
| LLY009 | 3.1 ± 0.3 | 2.37 ± 0.5 | |
| LLY010 | 0.3 ± 0.04 | 0.11 ± 0.01 | |
| LLY011 | 0.5 ± 0.1 | 0.08 ± 0.03 | |
| LLY012 | 0.25 ± 0.04 | 0.75 ± 0.1 | |
| LLY013 | 0.21 ± 0.05 | 0.4 ± 0.01 | |
| LLY014 | 0.29 ± 0.05 | 0.21 ± 0.03 | |
| LLY015 | 9.75 ± 1.23 | 12.1 ± 3.16 | |
| LLY016 | 13.8 ± 2.8 | 24.4 ± 2.39 |
Compounds were tested in rabbit plasma (rPlasma), rabbit caseum (rCaseum) and the surrogate matrix produced from human THP1 macrophages. RED assays were conducted in triplicates and fu is expressed as the mean ± SD.
Assays that had <70% recovery.
In Vitro Generation of a Caseum Surrogate
On the basis of the apparent correlation between the unbound drug fraction in caseum and passive diffusion into caseum in vivo, we set out to develop a medium throughput assay in order to expand the data set and explore relationships between caseum binding and physicochemical properties. To obviate the need for collecting caseum from infected animals, we designed a caseum mimic that would reproduce the binding properties of the actual matrix. THP-1 monocytes were exposed to 200– 1000 μM oleic acid (OA), which is known to induce differentiation into FM in vitro,18 and lipid droplet accumulation was detected by flow cytometry. PMA-differentiated THP-1 macrophages (THPM) accumulated lipid droplets most significantly in the presence of 400 μM OA (Figure 2a). Under these conditions, there was an 11-fold increase (from 9.7 × 105 to 1.1 × 106 MFI) in BODIPY 493/503-staining over control cells. Lower concentrations of OA resulted in poorer lipid droplet accumulation, whereas concentrations higher than 400 μM resulted in a decline in the florescence signal that can be attributed to the loss of cell viability and adhesion. We also determined that the recovery of lipid-loaded macrophages reached a peak after 24 h of OA exposure (Figure 2b). THPMs were treated with 400 μM OA for 24 h, and the resulting FMs were washed, harvested, lysed, and denatured prior to being used in the RED assay. The fractions unbound of 36 anti-TB drugs, metabolites, and noncommercial compounds in the caseum surrogate are listed in Table 1. Compound binding in the surrogate matrix correlated well with binding to native caseum, with the exception of ethionamide and pyrazinamide, which bind more extensively to surrogate (fu = 27.8 and 62%, respectively) than actual caseum (fu = 52.8 and >99.9%, respectively). The correlation coefficient R2 was 0.88 when all compounds were included and 0.95 after exclusion of the two outliers (Figure 2c).
Figure 2.
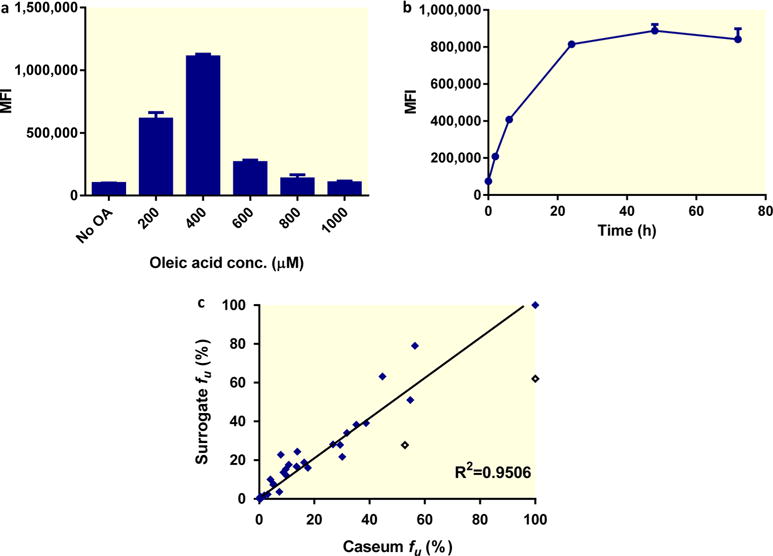
In vitro generation and performance of caseum surrogate. (a) Dose–response of lipid droplet accumulation in THPMs. Macrophages were exposed to increasing concentrations of oleic acid for 24 h, and lipid droplet accumulation was quantified by staining with BODIPY 493/503 and detection by flow cytometry. (b) Time-course of lipid droplet accumulation in THPMs exposed to 400 μM of oleic acid. Lipid droplet content was quantified using BODIPY and is presented as the mean fluorescence intensity (MFI). The average and standard deviations (error bars) of three independent experiments are shown. (c) Correlation between the fraction unbound (fu) of selected drugs in caseum and the surrogate matrix. The best-fit line excluding the outliers (hollow diamonds) was determined using linear regression. The goodness of fit is expressed as the R2 value.
Comparative Protein and Lipid Contents of Surrogate and Actual Caseum
To characterize the surrogate matrix and facilitate the interpretation of binding data, we measured the protein, lipid, and nucleic acid contents of rabbit plasma, caseum, and the surrogate matrix produced from FMs. We measured cholesterol and triglycerides because they represent the major lipid species found in caseum.15 Table 2 summarizes the average concentrations of these molecular species in rabbit plasma and in three independent batches of caseum and surrogate material. The protein content of plasma and caseum samples was 10-fold higher than in the surrogate matrix (54.7 μg/μL in plasma, 56.7 μg/mg in caseum, and 5.3 μg/mg in the surrogate). Combined tri-, di-, and monoglyceride concentrations were markedly higher in the surrogate (12.2 μg/mg) than in caseum (0.32 μg/mg) and plasma (0.21 μg/μL). Free cholesterol concentrations were comparable in caseum and the surrogate (3.19 and 2.82 μg/mg respectively) and around 20-fold higher than in rabbit plasma. RNA levels were similar in native caseum and in the surrogate (0.4 and 0.33 μg/mg, respectively), whereas the DNA content of caseum was approximately 10-fold lower than measured in the surrogate (0.17 and 2.0 μg/mg, respectively). To determine the variability in caseum and surrogate binding between different batches of both matrices, caseum samples from different rabbits and independent rounds of infection were used. Similarly, surrogate matrix samples produced from different batches of foamy THP-1 macrophages were compared. The fraction unbound of moxifloxacin was determined in both matrixes with triplicate samples for each batch (Figure 3a,b). In caseum, the mean fu for moxifloxacin ranged from 13.5 to 17.8%, and in the surrogate it ranged from 16.8 to 24.3%. Using one-way analysis of variance (ANOVA), we concluded that there is no batch effect at the 0.05 level of significance for either matrix in the RED assay.
Table 2.
Average Concentrations of Proteins, Free Cholesterol, Triglycerides, DNA, and RNA in Rabbit Plasma, Caseum, and Surrogate Caseum Produced from THPMa
| plasma (μg/μL) | caseum (μg/mg) | surrogate (μg/mg) | |
|---|---|---|---|
| protein | 54.7 ± 5.0 | 56.7 ± 4.7 | 5.3 ± 2.4 |
| free cholesterol | 0.14 ± 0.02 | 3.19 ± 0.72 | 2.82 ± 1.03 |
| triglycerideb | 0.21 ± 0.04 | 0.32 ± 0.02 | 12.2 ± 2.1 |
| DNA | 0.17 ± 0.01 | 2.0 ± 0.3 | |
| RNA | 0.40 ± 0.01 | 0.33 ± 0.1 |
Average values and standard deviations from three independent biological replicates are shown.
The commercial assay is provided as a method to quantify triglycerides but the read-out also includes di- and monoglycerides.
Figure 3.
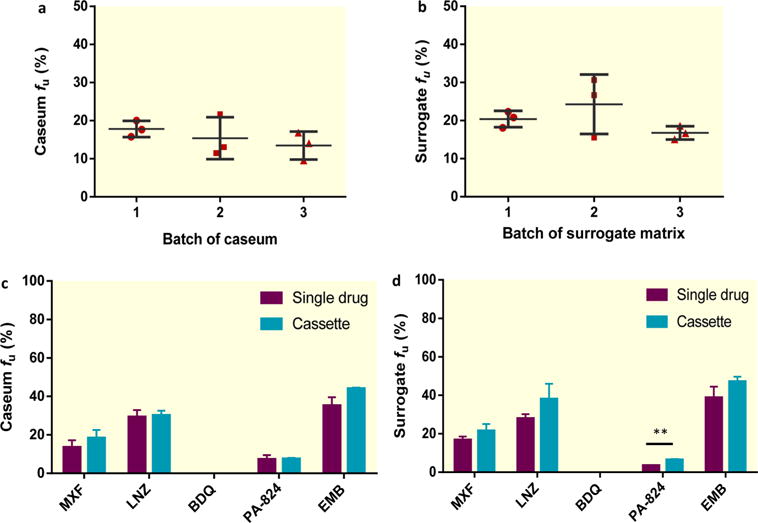
Reproducibility of the native and surrogate caseum assays. Analysis of batch effect on caseum (a) and surrogate matrix (b) binding of moxifloxacin (MXF). Dot plots, each with a mean and a standard deviation of three independent experiments, are shown. (c, d) Comparison between the fraction unbound (fu) of MXF, linezolid (LNZ), bedaquiline (BDQ), PA-824, and ethambutol (EMB) in (c) caseum and (d) the surrogate when tested individually (purple bars) and in combination (cassette testing) (blue bars). The average and standard deviations (error bars) of three independent experiments are shown. The statistically significant difference in fu is indicated as **, p < 0.01.
To increase throughput, we simultaneously determined the fraction unbound of five anti-TB drugs in caseum and the surrogate in a cassette format. We chose moxifloxacin, linezolid, bedaquiline, PA-824, and ethambutol because they span the full range of the extent of caseum binding. In caseum, no significant difference in binding was detected for any drug when comparing a single drug with the cassette assay format (Figure 3c). In the surrogate matrix, there was a general trend toward slightly decreased binding (increased fu) for all compounds in the cassette test, although only PA-824 showed a statistically significant difference (fu increased from 3.6 to 6.5%) (p < 0.01) (Figure 3d). Despite absolute differences in drug binding between the single drug and cassette formats, the overall ranking of compounds based on their fraction unbound (ethambutol > linezolid > moxifloxacin > PA-824 > bedaquiline) was maintained in both caseum and the surrogate matrix. By manipulating the protein and lipid contents of the caseum surrogate, we also determined that both proteins and lipids, as found in true caseum and the THP-1 lysate surrogate, were required to achieve full binding of all drugs tested (Figure S2b).
Physicochemical and Molecular Correlates of Caseum Binding
A superficial analysis of the association between physicochemical parameters and caseum binding revealed a weak correlation between hydrophobicity (calculated octanol/water partitioning coefficient, clogP) and binding to caseum, which is not surprising because lipophilic drugs generally exhibit higher nonspecific binding to macromolecules.19 To develop an in silico tool predictive of caseum binding, we profiled a large compound set including 213 small drug-like TB actives representing a variety of chemotypes and 30 compounds containing the pyrazinamide scaffold (Table S1). The set of compounds with anti-TB activity was derived from TB drug discovery efforts from various pharmaceutical and academic entities. The compounds with the pyrazinamide core were selected to cover a representative range of molecular weight (MW), clogP, hydrogen bond donor/acceptor groups, and ionizable groups. The fractions unbound of these compounds are listed in Table S1 and range between 0.5 and >99.9% (Figure S3), showing that binding can vary significantly between structurally related molecules that span a wide chemical space.
In silico descriptors were calculated for all compounds evaluated in the surrogate binding assay. Principal component analysis (PCA) identified 64 molecular descriptors that together described the fraction unbound in the caseum surrogate (fu surrogate). A two-dimensional PCA p1 versus p2 loading plot was generated to rank these descriptors on the basis of their distance to the fu surrogate in the p1/p2 space. Positive correlations were determined on the basis of the proximity to fu surrogate, whereas negative correlations were extrapolated from the projection of fu surrogate across the origin (Figure 4a). The minimum distance (d) observed (strongest correlation) was 0.022, with a total of 18 descriptors showing a distance <0.1. The top 13 descriptors are listed in Table 3, and selected individual correlations are illustrated in Figure S4a.
Figure 4.
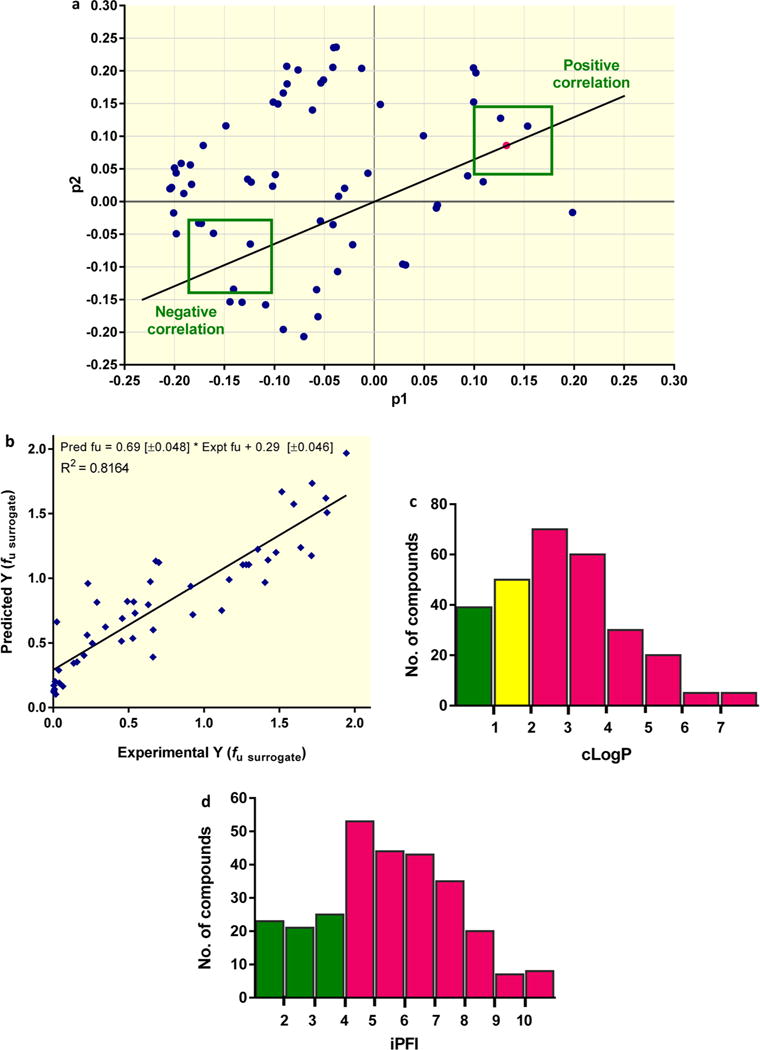
In silico predictive model of caseum binding. (a) The PCA loading p1 versus p2 plot shows how the different descriptors (blue dots) analyzed vary in relation to the experimental data (magenta dot) and to each other. The top 13 descriptors exhibiting the strongest correlation (negative or positive) are presented in Table 3. (b) The correlation between predicted fraction unbound (fu) and experimental fu data in surrogate caseum for all 49 compounds in the test set. The best-fit line was determined using linear regression. The goodness of fit is expressed as the R2 value. (c, d) Proportion (%) of compounds achieving fu > 10% in caseum surrogate with the 279 compounds categorized by (c) clogP or (d) ciPFI bins. The bars show the absolute number of compounds in each bin. Color coding refers to the % chance of achieving the benchmark value (fu > 10%) in that bin/category: green, > 67%; yellow, 34–67%; and magenta, <33%.
Table 3.
Top 13 Physicochemical and Molecular Descriptor Parameters That Correlate with the Fraction Unbound in the Caseum Surrogatea
| parameter | description | binning of parameter | correlation | 2D distance score |
|---|---|---|---|---|
| AromRings | aromatic rings | molecular shape character | − | 0.022 |
| Mol_Sol | molecular solubility | solubility | + | 0.036 |
| sp2 | sp2 carbon count | molecular shape character | − | 0.046 |
| D1 | hydrophobic volume | lipophilicity | − | 0.049 |
| IW1 | hydrophilicity/lipophilicity unbalance | lipophilicity | + | 0.061 |
| ID1 | hydrophilicity/lipophilicity unbalance | lipophilicity | + | 0.061 |
| Ring_count | ring count | molecular shape character | − | 0.066 |
| R | volume/surface ratio | molecular shape character | − | 0.068 |
| Logp_nOct | logP | lipophilicity | − | 0.068 |
| PB | plasma protein binding | plasma protein binding | − | 0.068 |
| CW2 | hydrophilic volume per surface unit | lipophilicity | + | 0.074 |
| HAS | hydrophobic surface area | lipophilicity | − | 0.075 |
| LogD | logD | lipophilicity | − | 0.076 |
The ranking of all correlations is based on the p1 and p2 PCA loading. The correlation is quantified by the distance between fu surrogate experimental data and the descriptor in the p1/p2 space (i.e., the shorter the distance, the greater the correlation). Positive correlations (i.e., fu increases with an increase in the parameter) and negative correlations (i.e., fu decreases with an increase in the parameter) are indicated by + and −, respectively.
The PCA loadings highlight how the experimentally derived fu surrogate clearly correlates with molecular solubility (d = 0.036), plasma protein binding (d = 0.069), the volume of the distribution (d = 0.089), and a number of molecular descriptors related to the lipophilic/hydrophilic character of the compounds (i.e., hydrophobic volume [d = 0.049], clogP [d = 0.068]) and their molecular size/shape (i.e., number of aromatic rings [d = 0.022], number of sp2 hybridized carbon atoms [d = 0.047], and volume/surface ratio [d = 0.068]).
The t1 versus t2 plot highlights how the set of 279 compounds provides reasonable coverage of the molecular property space as defined by the descriptors under evaluation (Figure S4b). We identified a number of outliers that are compounds that fall into the extreme boundaries of the molecular property space. They were either large, natural-product-like (rifampicin and rifapentine), very lipophilic (e.g., bedaquiline), highly polar–charged, or small fragment-like compounds (isoniazid, p-aminosalicylic acid).
In conclusion, the PCA analysis revealed high lipophilicity and poor solubility as positive drivers of binding to caseum. The negative correlation between the free fraction and the number of aromatic rings, number of sp2 carbon counts, and volume-to-surface ratio indicates that low-molecular-weight nonflat compounds are more likely to exhibit low caseum binding properties.
In Silico Predictive Model of Caseum Binding
The 279 compound data set was used to develop a statistical quantitative structure–activity relationship model for the prediction of the caseum binding of new compounds. The set of 279 compounds was randomly divided so that a training set (80% of the total) could be used for model learning to predict the free-energy-relationship-related response variable Y (Materials and Methods) for the test set (20% of the total). To assess the robustness and utility of the consensus model as a predictive tool, the compounds in the test set were evaluated and a goodness of fit of R2 = 0.82 was obtained between the predicted and experimental values (Figure 4b). The average structural similarity (defined as the Tanimoto similarity (T) index using extended connectivity fingerprints ECFP6 as the molecular descriptor)20,21 of the compounds in the test set compared to the training set was T = 0.44 (Tmin = 0.11 and Tmax = 0.80, with T = 1 for the highest similarity and T = 0 for lowest similarity).
Having a robust in silico predictive model relying on a large number of molecular descriptors, we next explored the possibility of using a very small set of physicochemical descriptors as an intuitive guide during the early stages of TB drug discovery. Composite measures of hydrophobicity/lipophilicity and aromatic ring count have been projected as simple property forecast indices. Specifically, the intrinsic property forecast index iPFI, defined as the sum of the experimental logP and number of aromatic rings (iPFI = logP + #Ar),22 is used by medicinal chemists to assess the quality of property-based designs in hit-to-lead optimization phases. Because iPFI includes two descriptors that ranked high in our PCA analysis (Table 2), we evaluated its predictive value as a simplified caseum forecast index. For this purpose, we considered a slightly modified composite property, ciPFI [clogP + #Ar], based on calculated logP rather than experimental logP. ciPFI showed a stronger correlation with fu surrogate (R2 = 0.45) (Figure S4c) than the number of aromatic rings (R2 = 0.25) or logP (R2 = 0.41) alone (Figure S4a), confirming that the composite index could be useful in TB drug discovery. On the basis of the observed in vivo partitioning of TB drugs at the interface between the cellular rim and the necrotic core of lung lesions (Figure 1), we set a threshold of fu = 10% to evaluate the predictive power of ciPFI [clogP + #Ar] versus clogP (Figure 4c,d). The results showed that compounds with clogP < 1 had a high chance (>67%) of achieving fu > 10%. Compounds with ciPFI < 4 had a high chance of achieving the same benchmark.
DISCUSSION
The site of many infectious diseases is outside the blood or plasma compartment. In TB disease, the pathogen resides in lesions that often contain a necrotic core devoid of a vascular supply, resulting in poor diffusion of some drugs but not others.2 The discovery of more effective anti-TB drugs requires a deeper understanding of the drivers of drug penetration in the caseum of TB lesions because necrotic granulomas and cavities, both containing caseum, are difficult to sterilize.9–11 We have previously shown, by MALDI mass spectrometry imaging (MSI) and the conventional quantitation of drugs in TB lesions, that different drugs display very distinct partitioning into the cellular and caseous regions of granulomas and cavities.2,13,17 Because necrotic material or caseum is entirely acellular and thus devoid of a vascular supply and active transport, only the free drug fraction can penetrate this matrix via passive diffusion. We hypothesized that factors influencing passive diffusion into caseum would include intrinsic physicochemical properties and the extent of binding to caseum macromolecules. As drug molecules diffuse inward from the cellular and vascularized regions of the granuloma, they bind to macromolecules at the outer edge of the caseous core, which prevents further passive diffusion toward the center of the necrotic core. To confirm this hypothesis, we developed an in vitro assay of caseum binding and showed that passive drug diffusion through caseum in vivo increases as the fraction unbound of drug in caseum ex vivo increases (Figure 1). By adapting the caseum binding assay to ensure medium throughput, we have generated a sizable data set from which a robust computational model was built to predict caseum binding from 64 physicochemical and molecular descriptors.
The rapid equilibrium dialysis methodology was used to measure drug binding in small volumes of caseum because it has been validated with various tissue homogenates.23 Mass balance was systematically included to rule out nonspecific binding of compounds to the device. A caseum surrogate was developed and optimized to fulfill two major objectives: (1) work with a noninfectious matrix (caseum harbors high mycobacterial loads) and (2) increase the assay throughput by circumventing the need for infecting animals. The method developed to generate the mimic was based on the premise that caseum arises from the necrosis/apoptosis of foamy macrophages, among other cell types, in the center of mature granulomas. In biopsy samples from patients with sputum-positive TB, foamy macrophages are found only in necrotic lesions, suggesting that they serve an active function in necrosis formation and accumulation of caseous debris at the heart of the granuloma.24,25 The linear correlation (R2 = 0.88 to 0.95) observed over the full range of binding in actual caseum versus the surrogate matrix confirmed our assumption that oleic acid-induced and -lysed foamy macrophages reasonably mimic caseum (Figure 2c). The pH of the synthetic caseum was neutral (data not shown), as has been shown for native caseum by several groups.26,27 Modifications to the caseum mimic revealed that denatured proteins and lipids both contribute to the overall caseum binding capacity (Figure S2b). To increase assay throughput, compounds can be tested using a cassette format in the caseum surrogate (Figure 3d), and the use of an easily detectable medium-binding compound (i.e., moxifloxacin) as an internal standard in every cassette assay allows for monitoring the performance of the assay and the matrix.
Of the initial set of 36 compounds that were tested in true caseum and in the surrogate, pyrazinamide and ethionamide, both containing a primary amide, stood as outliers with lower fractions unbound in the surrogate matrix than in native caseum. Each assay was performed in triplicate (with three separate batches of matrix in the case of pyrazinamide) with good reproducibility, showing a significant difference in binding between the mimic and true caseum (p = 0.009 and 0.001 for pyrazinamide and ethionamide, respectively). A component present in the surrogate matrix but not in caseum appears to be responsible for the significant binding of pyrazinamide and ethionamide. Published human plasma protein binding of pyrazinamide varies between 10 and 50%,28–30 suggesting that the drug is highly susceptible to subtle differences in matrix composition. Small and polar molecules other than pyrazinamide and ethionamide exhibited a good binding correlation between the two matrixes.
PCA integrating 64 physicochemical and molecular parameters identified the strongest correlation between caseum binding and the number of aromatic rings (2D distance score of 0.022), followed by solubility, the number of sp2 carbons, and various lipophilicity parameters (Figure 4a). The most influential parameters (2D distance score of <0.08) were binned, revealing a high predictive value for molecular shape, lipophilicity, solubility, and plasma protein binding (Table 3). Plasma protein binding alone, however, did not constitute an acceptable surrogate of caseum binding (Figure S1). The pKa was not detected as a driver of binding to caseum macromolecules and is thus not expected to influence passive diffusion through this matrix, the pH of which is in the neutral range.27,31 The individual parameters highlighted in Figure S4a appear to weakly correlate with surrogate fu on the basis of their low goodness-of-fit scores (0.05 < R2 < 0.41). The response variables are positioned in a multidimensional descriptor space defined by the selected descriptor set. The PCA statistical procedure defines a low-dimensional projection hyperplane in this space, oriented to maximize the variance of the response variable data. Hence, it is no surprise that individual descriptors show a weaker correlation with the response variable data than the principal components that span this information-rich hyperplane. Using 69 calculated molecular descriptors, a consensus computational predictive model was built from 279 compounds randomly divided into training (80%) and test (20%) sets, with a goodness of fit of R2 = 0.82, suggestive of a robust in silico predictive model (Figure 4b). Because the PCA signature was derived from a data set of binding to caseum surrogate, we built a test set with 10 compounds for which true caseum binding data was available. For these 10 compounds, the predicted Y = log(1 + X) value correlates well with an R2 of 0.92 (not shown), thus validating the molecular signature as representative of true caseum binding.
Composite measures of hydrophobicity/lipophilicity and the aromatic ring count have been projected as simple property forecast indices (e.g., the intrinsic property forecast index iPFI defined as the sum of the chromatographically determined logP and the number of aromatic rings, iPFI = Chrom logP + #Ar)22 and are used by medicinal chemists to assess the quality of property-based designs in hit-to-lead optimization phases. Because iPFI relates to key physicochemical and molecular descriptors that ranked high in our PCA analysis (Table 3), we were interested in evaluating its predictive value as a simplified caseum forecast index. For this purpose, we considered a slightly modified composite property, ciPFI [clogP + #Ar], based on the calculated logP rather than Chrom logP. We concluded that the summing of hydrophobicity value plus aromatic ring count provides a simple caseum forecast tool for TB drug discovery. Together, the consensus computational predictive model and/or the ciPFI are useful in guiding the property-based design in the hit-to-lead optimization phases of TB drug discovery.
Despite the fact that most compounds had anti-TB activity, the molecular property space as defined by the 64 descriptors under evaluation was well covered (Figure S4b). This is likely related to the extreme diversity of the chemical space of TB actives.32 We propose to deploy the prediction model as a high-throughput classification tool to categorize low, medium, and high fraction unbound compounds in TB drug discovery programs rather than predict the exact value of the free fraction. This model is well suited to building focused compound libraries for TB screening programs and to increasing the probability of identifying TB drug candidates with desirable lesion penetration.
A limitation of this study is the small number of TB drugs for which MALDI images of partitioning in necrotic lesions are available. Although a clear association is observed between caseum binding and inward diffusion toward the center of caseous foci (Figure 1), additional data points are required to strengthen the inverse correlation between passive diffusion and binding to caseum components. Strength of binding to caseum macromolecules and active uptake into the foamy macrophage layer that directly subtends the necrotic center, including lysosomal trapping, are also likely to influence drug partitioning.2
Given the heterogeneous nature of TB lesions and the differential drug susceptibility of bacterial subpopulations within these environments, combination therapies are necessary for the effective treatment of TB. However, current drug combinations have not taken into account the differential abilities of drugs to penetrate the various layers of multiple lesion types. The caseum binding assay, in silico consensus model, and simplified ciPFI signature described in this study are translational tools with which drug penetration into the caseous core of necrotic lesions can be predicted in a medium-throughput manner to guide discovery compound optimization, select focused compound libraries for screening programs, and guide the design of more effective combination regimens.
MATERIALS AND METHODS
Antimicrobials and Reagents
Moxifloxacin and linezolid were purchased from Sequoia Research Products (Pangbourne, U.K.). Levofloxacin, pyrazinamide, clofazimine, isoniazid, rifapentine, para-aminosalicylic acid (PAS), and ethionamide were purchased from Sigma-Aldrich (St. Louis, MO, USA). Acetyl-isoniazid was purchased from Santa Cruz Biotechnology (Dallas, TX, USA). Ethambutol was purchased from Alfa Aesar (Ward Hill, MA, USA). Rifampicin was purchased from Gold Biotechnology (Olivette, MO, USA). Gatifloxacin was purchased from Chem-Impex International Inc. (Bensenville, IL, USA). Posizolid, radezolid, and tedizolid were kindly provided by Astra-Zeneca, Rib-X, and Trius Pharmaceuticals, respectively. OPC67683 (delamanid) and sutezolid were synthesized as described below. PA-824 was provided by the Global Alliance for TB Drug Development (New York, NY, USA), and bedaquiline was provided by Janssen Research and Development (Titusville, NJ, USA). Eli Lilly and Co. (Indianapolis, IN, USA) kindly provided a set of 16 noncommercial compounds of undefined bactericidal activity in order to expand the range of physicochemical properties. Additional sets of 30 pyrazinamide-containing compounds and 213 drug-like small molecules with anti-TB activity, derived from TB drug discovery efforts carried out as a collaboration among the NIH, the University of Dundee (the HIT-TB program), and the TB Drug Accelerator Program of the Bill and Melinda Gates Foundation (http://partnerships.ifpma.org/partnership/tb-drug-accelerator-program), were provided by the various partners. Female rabbit plasma (K2 EDTA) was purchased from Bioreclamation IVT (Hicksville, NY, USA).
Synthesis of Selected TB Drugs
Sutezolid and its metabolites were synthesized according to published procedures,33 with a modified purification procedure for sutezolid. Following the Staudinger reduction and acylation, the crude reaction mixture was concentrated in vacuo using toluene to azeotrope the remaining pyridine. The crude reaction mixture was then adsorbed to silica and FCC in 10–30% acetone/DCM. On larger scales, sutezolid was further purified using reverse-phase prep-HPLC (Varian Prostar, model 218 pumps, model 325 UV detector monitoring at 254 nm, model 701 fraction collector, on a Phenomenex Luna C18 column, 100 Å, 250 × 21.20 mm2). The compounds were eluted over a 24 min gradient from 18 to 65% acetonitrile to remove trace triphenylphosphine oxide. The resultant solid was recrystallized from ethanol to give a white or pale-pink crystalline solid.
OPC67683 was synthesized according to published procedures with slight modifications to the procedure.34 The Sharpless asymmetric epoxidation was performed using (D)-(−)-diethyl tartrate rather than (D)-(−)-diisopropyl tartrate. Purification of the final compound was effected by column chromatography on silica gel in 1:9 MeOH/DCM (v/v) followed by recrystallization from ethanol to give the compound as a very pale yellow solid.
Rabbit Infection Model and Caseum Collection
New Zealand White (NZW) rabbits were used for aerosol infection by M. tuberculosis HN878 or M. bovis AF2122, as previously described.35,36 Briefly, rabbits were exposed to M. tuberculosis-or M. bovis-containing aerosol using a nose-only delivery system. The infection was allowed to progress for 4 weeks (M. bovis) or 12 to 16 weeks (M. tuberculosis) prior to necropsy and the collection of caseum from both closed and cavitary lesions and was used indiscriminately. Possible differences between nodule caseum and cavity caseum include the oxygen tension (lower in closed nodule caseum) and cellularity (potentially higher in cavity caseum). However, live cells present in cavity caseum were unlikely to survive the freeze–thaw cycle and should not have interfered with the assay. Animal studies were carried out in accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health, with approval from the Institutional Animal Care and Use Committee of the New Jersey Medical School, Newark, NJ, and the NIAID (NIH), Bethesda, MD.
Generation of Foamy Macrophages and Surrogate Caseum
The THP-1 monocytic cell line was cultured in RPMI 1640 supplemented with 10% FBS and 2 mM L-glutamine. Cells were seeded on plates (1 × 106 cells/mL), exposed to 100 nM phorbol 12-myristate 13-acetate, and allowed to adhere. To simulate caseum formation, lipid body production in THP-1 monocytes was stimulated overnight with 0.4 mM oleic acid (OA), which is known to induce differentiation into FM in vitro.18 FMs were detached, washed three times with phosphate-buffered saline (PBS), and pelleted. Cells were lysed during three free–thaw cycles, and proteins were denatured by a 30 min incubation period at 75 °C to mimic alterations in protein structure and 3D conformation that occur in the necrotic core of granulomas.
The protocol was also manipulated to produce variations of the surrogate matrix. THP-1 macrophages were simply detached and harvested after PMA stimulation if no oleic acid stimulation was required. A test surrogate matrix was also produced, having omitted the denaturation period at 75 °C. Lipid extracts of the surrogate matrix were derived by extracting it 10:1 (v/w) with a 2:1 chloroform/methanol mixture.
Flow Cytometry
Lipid accumulation in FMs was detected using immunofluorescence flow cytometric analysis. Foamy macrophages were fixed in 4% paraformaldehyde (in PBS) for 30 min and then washed and blocked with 5% bovine serum albumin (BSA). The neutral lipids in lipid bodies were stained with 10 μg/mL BODIPY 493/503 (Molecular Probes) for 30 min at room temperature in the dark. The stained cells were washed three times with 5% BSA and stored at 4 °C. Stained cells were detected using Accuri C6, and data analysis was performed using the CFlow Plus software (BD Biosciences). Up to 50 000 events were detected per sample. Events were gated on the forward and side scatters to select for the main population of cells. The mean fluorescence intensity (MFI) for the 533/30 emission channel was generated for the gated events.
Rapid Equilibrium Dialysis (RED) Assay
All binding assays were conducted using the disposable RED device (ThermoFisher Scientific, Waltham, MA), which is more commonly used to measure plasma protein binding. Previous studies have shown that a 4 h time course is sufficient for the equilibration of a variety of compounds between chambers.37 Caseum and the caseum surrogate were diluted 10-fold in PBS and homogenized. Plasma/caseum/surrogate was spiked to give the final incubation concentration of 5 μM. Spiked plasma (200 μL) was placed in the sample chambers, and the buffer chambers were filled with 350 μL of PBS. The plates were then covered with adhesive seals and incubated at 37 °C for 4 h on an orbital shaker set at 300 rpm. Following incubation, samples were removed from both chambers and processed by adding an organic solvent mixture (1:1 methanol/acetonitrile) prior to LC-MS quantification. The fractions unbound were calculated using methods discussed in previous studies.38,39 The fraction unbound (fu) in plasma and diluted caseum/surrogate was calculated as the ratio between free (buffer chamber) and total drug (sample chamber) concentrations, as shown in eq 1. A dilution factor of 10 (D = 10) was applied to the calculation of fu in undiluted caseum and surrogate, as shown in eq 2.39 The recovery for each assay was calculated using eq 3 and was considered to be acceptable between 70 and 130%.38 Two-tailed t tests were used to compare binding to native and surrogate caseum.
| (1) |
| (2) |
| (3) |
Surrogate caseum binding assays that were run in cassette format included five test compounds at 5 μM. All cassette assays also included moxifloxacin to ensure consistency between batches. When lipid extracts were used in the binding assay, both caseum and the surrogate matrix were extracted with a 2:1 chloroform/methanol mixture. The supernatants were air dried, and the residual lipids were resuspended in PBS by sonicating in a water bath.
Quantitative Analysis
Drug quantification was achieved by using liquid chromatography coupled to mass spectrometry (LC-MS). Mass analysis and detection were performed on an Applied Biosystems (Manchester, U.K.) 4000 Q-trap triple-quadrupole mass spectrometer equipped with a turbo ion-spray ionization source. The HPLC system is of the Agilent (Santa Clara, CA) 1260 series with a degasser, binary pump, autosampler, and thermostated column compartment. Chromatographic separation for all compounds was achieved with an Agilent Zorbax SB-C8 (2.1 × 30 mm2, 3.5 μm) column using gradient conditions unless otherwise mentioned. PAS, ethionamide, pyrazinamide, and the pyrazinamide-containing compound set provided by the Drug Discovery Unit of Dundee University were analyzed using the Agilent Zorbax SB-C8 (4.6 × 75 mm2 3.5 μm) column. The analysis of ethambutol and isoniazid required normal phase methods on a Cogent Diamond Hydride column (2.1 × 50 mm2, 4 μm) (MicroSolv Technology Corp, Eatontown, NJ). Mobile phases, water and acetonitrile, were acidified with 0.1% acetic acid. Data collection and processing were performed with Analyst 1.6.2 software. Responses from sample chambers and buffer chambers were normalized to internal standard (IS) peak areas. Stably labeled isotopes isoniazid-D4 (C/D/N Isotopes, Quebec, Canada), acetyl-isoniazid-D4 (Toronto Research Chemicals, Ontario, Canada), pyrazinamide-15N-D3 (Sigma-Aldrich), pyrazinoic acid-D3 (Toronto Research Chemicals), and ethambutol-D10 (C/D/N Isotopes) were used as internal standards for isoniazid, acetyl-isoniazid, pyrazinamide, pyrazinoic acid, and ethambutol, respectively. Diclofenac or verapamil was used for all other compounds.
Protein, Lipid, and Nucleic Acid Quantification
The total protein content was determined using a Coomassie (Bradford) protein assay kit (Thermo Scientific). Triglyceride and cholesterol quantification was achieved using colorimetric kits from BioVision (Milpitas, CA, USA). Note that the method used for triglyceride quantification also captures di- and monoglycerides (http://www.biovision.com/manuals/K622.pdf). RNA and DNA isolation from caseum and the surrogate was achieved using the RNeasy Mini Kit and QiaAMP Fast DNA Tissue Kit from Qiagen (Valencia, CA), respectively. DNA and RNA concentrations were quantified using a Nanodrop 2000 spectrophotometer (Thermo Scientific).
Data Analysis and Computational Modeling
A set of 64 in-silico descriptors were calculated for all compounds evaluated in the surrogate matrix binding assay. Parameters directly related to the compounds’ molecular properties (e.g., MW, number of rotatable bonds, number of hydrogen bond donors and acceptors, number of heavy atoms, number of sp3-and sp2-hybridized carbon atoms, polar molecular surface area, and molecular volume) were generated in Pipeline Pilot (PipelinePilot version 9.1.0.13 by BIOVIA, http://accelrys.com/products/collaborative-science/biovia-pipeline-pilot/), Stardrop (version 6.0 by Optibrium, www.optibrium.com/stardrop), and Volsurf+ (version 1.0.7.l by Molecular Discovery, www.moldiscovery.com/software/vsplus).40,41 An additional set of descriptors based on 3D maps of molecular interaction fields were also generated using Volsurf+. Within Volsurf+, the area of the molecule that can establish favorable interactions with a number of chemical probes is mapped for each compound. From this unique molecular representation, Volsurf+ derives a number of descriptors that proved to be relevant in modeling compounds’ ADME properties. For each compound, the molecular solubility, clogP, plasma protein binding, and volume of distribution were also calculated using Pipeline Pilot/Volsurf+. In addition, PipelinePilot descriptors such as circular structural feature fingerprints [functional-class fingerprints (FCFP12), Sybyl atom-type extended connectivity fingerprints (SCFP4),21 E-state fragment keys,42 and spiro atom counts were used to take into account the molecular structure and the atomic electronic properties of the compounds. The complete list of calculated descriptors and their definition can be found in Table S3.
To assess the correlation between the different descriptors and surrogate matrix binding data, principal component analysis (PCA-XY, eight components, R2cum = 0.803, Q2cum = 0.645) was carried out using multivariate data analysis package Simca (Simca version 14.0.0.1359 by Umetrics, http://umetrics.com/products/simca). A computational predictive model was built using more than 150 molecular descriptors from which a subset of 69 properties were retained. To increase the robustness of the predictive computational model tool, a consensus approach was implemented combining a total of four different models within the Pipeline Pilot platform. These models are based on (1) a partial least-squares projection to latent structures regression (PLS) and (2) three different supervised machine learning methods [decision tree ensemble (DT), support vector machine regression (SVM), and artificial neural network (NN)]. The set of 279 compounds was randomly divided into training (230 compounds, 80% of the total) and test (49 compounds, 20% of the total) sets. The training set was used for model learning to predict the free-energy-relationship-related response variable Y = log (1 + X), where X = fu in the caseum surrogate, for each new compound (Y = 2 corresponds to the 100% free fraction whereas Y = 1 corresponds to the 0% free fraction). The four values of Y generated by each individual model were then averaged to generate a final consensus prediction of Y. The Tanimoto index or Tanimoto coefficient of similarity for molecules A and B was calculated as SAB = c/(a + b − c) where a is the fingerprint bits in A, b is the fingerprint bits in B, and c is the number of fingerprint bits common to both molecules.20
Supplementary Material
Acknowledgments
We thank Johnson & Johnson, the TB Alliance, Astra Zeneca, Rib-X, and Trius Therapeutics for providing bedaquiline, PA-824 (pretomanid), AZD5847, radezolid, and tedizolid, respectively. Brendan Prideaux, Matthew Zimmerman, Stephen Juzwin, Emma Rey-Jurado, Nancy Ruel, Leyan Li, and Danielle Weiner provided support with MALDI analysis, bioanalytical methods, preparation of the caseum surrogate, chemical synthesis, and isolation of rabbit caseum. We thank the members of the TB Drug Accelerator consortium and the University of Dundee/NIH HIT-TB consortium for selecting and sharing the compound series. We are grateful to Peter Warner and James Metz for stimulating discussions. This work was conducted with funding from the Bill and Melinda Gates Foundation, award nos. OPP1044966 and OPP1024050 to V.D., NIH Shared Instrumentation Grant S10OD018072, as well as joint funding from the Bill and Melinda Gates Foundation and Wellcome Trust for A Centre of Excellence for Lead Optimization for Diseases of the Developing World to P.W.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsinfecdis.6b00051.
A complete list of the compounds and descriptors analyzed and additional supporting data (PDF)
Author Contributions
J.P.S. designed and carried out all in vitro assays and drafted the manuscript; F.Z. analyzed and modeled the data; H.H. assisted with the high-throughput binding assay; L.S. assisted with data analysis and model building; L.E.V. provided ex vivo caseum from infected rabbits; G.A.M. synthesized selected TB drug candidates; T.M., F.Z., P.R., and P.W. selected compounds with defined physicochemical properties and aided with writing the manuscript; and V.D. conceived the study and wrote the manuscript.
Notes
The authors declare no competing financial interest
References
- 1.Dartois V. The path of anti-tuberculosis drugs: from blood to lesions to mycobacterial cells. Nat Rev Microbiol. 2014;12:159–167. doi: 10.1038/nrmicro3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prideaux B, Via LE, Zimmerman MD, Eum S, Sarathy J, O’Brien P, Chen C, Kaya F, Weiner DM, Chen PY, Song T, Lee M, Shim TS, Cho JS, Kim W, Cho SN, Olivier KN, Barry CE, 3rd, Dartois V. The association between sterilizing activity and drug distribution into tuberculosis lesions. Nat Med. 2015;21:1223–1227. doi: 10.1038/nm.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sacchettini JC, Rubin EJ, Freundlich JS. Drugs versus bugs: in pursuit of the persistent predator Mycobacterium tuberculosis. Nat Rev Microbiol. 2008;6:41–52. doi: 10.1038/nrmicro1816. [DOI] [PubMed] [Google Scholar]
- 4.Hoff DR, Ryan GJ, Driver ER, Ssemakulu CC, De Groote MA, Basaraba RJ, Lenaerts AJ. Location of intra- and extracellular M. tuberculosis populations in lungs of mice and guinea pigs during disease progression and after drug treatment. PLoS One. 2011;6:e17550. doi: 10.1371/journal.pone.0017550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peyron P, Vaubourgeix J, Poquet Y, Levillain F, Botanch C, Bardou F, Daffe M, Emile JF, Marchou B, Cardona PJ, de Chastellier C, Altare F. Foamy macrophages from tuberculous patients’ granulomas constitute a nutrient-rich reservoir for M. tuberculosis persistence. PLoS Pathog. 2008;4:e1000204. doi: 10.1371/journal.ppat.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gillespie SH, Basu S, Dickens AL, O’Sullivan DM, McHugh TD. Effect of subinhibitory concentrations of ciprofloxacin on Mycobacterium fortuitum mutation rates. J Antimicrob Chemother. 2005;56:344–348. doi: 10.1093/jac/dki191. [DOI] [PubMed] [Google Scholar]
- 7.Pasipanodya JG, Gumbo T. A new evolutionary and pharmacokinetic-pharmacodynamic scenario for rapid emergence of resistance to single and multiple anti-tuberculosis drugs. Curr Opin Pharmacol. 2011;11:457–463. doi: 10.1016/j.coph.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lenaerts AJ, Hoff D, Aly S, Ehlers S, Andries K, Cantarero L, Orme IM, Basaraba RJ. Location of persisting mycobacteria in a Guinea pig model of tuberculosis revealed by r207910. Antimicrob Agents Chemother. 2007;51:3338–3345. doi: 10.1128/AAC.00276-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin PL, Dartois V, Johnston PJ, Janssen C, Via L, Goodwin MB, Klein E, Barry CE, 3rd, Flynn JL. Metronidazole prevents reactivation of latent Mycobacterium tuberculosis infection in macaques. Proc Natl Acad Sci U S A. 2012;109:14188–14193. doi: 10.1073/pnas.1121497109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Via LE, England K, Weiner DM, Schimel D, Zimmerman MD, Dayao E, Chen RY, Dodd LE, Richardson M, Robbins KK, Cai Y, Hammoud D, Herscovitch P, Dartois V, Flynn JL, Barry CE., 3rd A Sterilizing Tuberculosis Treatment Regimen Is Associated with Faster Clearance of Bacteria in Cavitary Lesions in Marmosets. Antimicrob Agents Chemother. 2015;59:4181–4189. doi: 10.1128/AAC.00115-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aber VR, Nunn AJ. Short term chemotherapy of tuberculosis. Factors affecting relapse following short term chemotherapy. Bull Int Union Tuberc. 1978;53:276–280. [PubMed] [Google Scholar]
- 12.Chang KC, Leung CC, Yew WW, Ho SC, Tam CM. A nested case-control study on treatment-related risk factors for early relapse of tuberculosis. Am J Respir Crit Care Med. 2004;170:1124–1130. doi: 10.1164/rccm.200407-905OC. [DOI] [PubMed] [Google Scholar]
- 13.Kjellsson MC, Via LE, Goh A, Weiner D, Low KM, Kern S, Pillai G, Barry CE, 3rd, Dartois V. Pharmacokinetic evaluation of the penetration of antituberculosis agents in rabbit pulmonary lesions. Antimicrob Agents Chemother. 2012;56:446–457. doi: 10.1128/AAC.05208-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prideaux B, Dartois V, Staab D, Weiner DM, Goh A, Via LE, Barry CE, 3rd, Stoeckli M. High-sensitivity MALDI-MRM-MS imaging of moxifloxacin distribution in tuberculosis-infected rabbit lungs and granulomatous lesions. Anal Chem. 2011;83:2112–2118. doi: 10.1021/ac1029049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim MJ, Wainwright HC, Locketz M, Bekker LG, Walther GB, Dittrich C, Visser A, Wang W, Hsu FF, Wiehart U, Tsenova L, Kaplan G, Russell DG. Caseation of human tuberculosis granulomas correlates with elevated host lipid metabolism. EMBO Mol Med. 2010;2:258–274. doi: 10.1002/emmm.201000079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Driver ER, Ryan GJ, Hoff DR, Irwin SM, Basaraba RJ, Kramnik I, Lenaerts AJ. Evaluation of a mouse model of necrotic granuloma formation using C3HeB/FeJ. mice for testing of drugs against Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2012;56:3181–3195. doi: 10.1128/AAC.00217-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Irwin SM, Prideaux B, Lyon ER, Zimmerman MD, Brooks EJ, Schrupp CA, Chen C, Reichlen MJ, Asay BC, Voskuil MI, Nuermberger EL, Andries K, Lyons MA, Dartois V, Lenaerts AJ. Bedaquiline and Pyrazinamide Treatment Responses Are Affected by Pulmonary Lesion Heterogeneity in Infected C3HeB/FeJ Mice. ACS Infect Dis. 2016;2:251–267. doi: 10.1021/acsinfecdis.5b00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.den Hartigh LJ, Connolly-Rohrbach JE, Fore S, Huser TR, Rutledge JC. Fatty acids from very low-density lipoprotein lipolysis products induce lipid droplet accumulation in human monocytes. J Immunol. 2010;184:3927–3936. doi: 10.4049/jimmunol.0903475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamazaki K, Kanaoka M. Computational prediction of the plasma protein-binding percent of diverse pharmaceutical compounds. J Pharm Sci. 2004;93:1480–1494. doi: 10.1002/jps.20059. [DOI] [PubMed] [Google Scholar]
- 20.Flower DR. On the Properties of Bit String-Based Measures of Chemical Similarity. J Chem Inf Comput Sci. 1998;38:379–386. [Google Scholar]
- 21.Rogers D, Hahn M. Extended-connectivity fingerprints. J Chem Inf Model. 2010;50:742–754. doi: 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- 22.Young RJ, Green DV, Luscombe CN, Hill AP. Getting physical in drug discovery II: the impact of chromatographic hydrophobicity measurements and aromaticity. Drug Discovery Today. 2011;16:822–830. doi: 10.1016/j.drudis.2011.06.001. [DOI] [PubMed] [Google Scholar]
- 23.Liu X, Van Natta K, Yeo H, Vilenski O, Weller PE, Worboys PD, Monshouwer M. Unbound drug concentration in brain homogenate and cerebral spinal fluid at steady state as a surrogate for unbound concentration in brain interstitial fluid. Drug Metab Dispos. 2009;37:787–793. doi: 10.1124/dmd.108.024125. [DOI] [PubMed] [Google Scholar]
- 24.Russell DG, Cardona PJ, Kim MJ, Allain S, Altare F. Foamy macrophages and the progression of the human tuberculosis granuloma. Nat Immunol. 2009;10:943–948. doi: 10.1038/ni.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guirado E, Schlesinger LS, Kaplan G. Macrophages in tuberculosis: friend or foe. Semin Immunopathol. 2013;35:563–583. doi: 10.1007/s00281-013-0388-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lanoix JP, Ioerger T, Ormond A, Kaya F, Sacchettini J, Dartois V, Nuermberger E. Selective Inactivity of Pyrazinamide against Tuberculosis in C3HeB/FeJ Mice Is Best Explained by Neutral pH of Caseum. Antimicrob Agents Chemother. 2016;60:735–743. doi: 10.1128/AAC.01370-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koller F, Leuthardt F. NEKROSE UND AUTOLYSE. BEITRAG ZUR KENNTNIS DER DYSTROPHI-SCHEN VERKALKUNG. Klin Wochenschr. 1934;13:1527–1529. [Google Scholar]
- 28.Gumbo T, Siyambalapitiyage Dona CS, Meek C, Leff R. Pharmacokinetics-Pharmacodynamics of Pyrazinamide In A Novel In Vitro Model of Tuberculosis for Sterilizing Effect: A Paradigm for Faster Assessment of New Antituberculosis Drugs. Antimicrob Agents Chemother. 2009;53:3197. doi: 10.1128/AAC.01681-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kiem S, Schentag JJ. Interpretation of antibiotic concentration ratios measured in epithelial lining fluid. Antimicrob Agents Chemother. 2008;52:24–36. doi: 10.1128/AAC.00133-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Woo J, Cheung W, Chan R, Chan HS, Cheng A, Chan K. In vitro protein binding characteristics of isoniazid, rifampicin, and pyrazinamide to whole plasma, albumin, and alpha-1-acid glycoprotein. Clin Biochem. 1996;29:175–177. doi: 10.1016/0009-9120(95)02024-1. [DOI] [PubMed] [Google Scholar]
- 31.Lanoix JP, Ioerger T, Ormond A, Kaya F, Sacchettini J, Dartois V, Nuermberger E. Selective inactivity of pyrazinamide against tuberculosis in C3HeB/FeJ mice is best explained by neutral pH of caseum. Antimicrob Agents Chemother. 2016;60:735. doi: 10.1128/AAC.01370-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koul A, Arnoult E, Lounis N, Guillemont J, Andries K. The challenge of new drug discovery for tuberculosis. Nature. 2011;469:483–490. doi: 10.1038/nature09657. [DOI] [PubMed] [Google Scholar]
- 33.Barbachyn MR, Hutchinson DK, Brickner SJ, Cynamon MH, Kilburn JO, Klemens SP, Glickman SE, Grega KC, Hendges SK, Toops DS, Ford CW, Zurenko GE. Identification of a novel oxazolidinone (U-100480) with potent antimycobacterial activity. J Med Chem. 1996;39:680–685. doi: 10.1021/jm950956y. [DOI] [PubMed] [Google Scholar]
- 34.Yamamoto A, Shinhama K, Aki S, Ogasawara S, Utsumi N. Synthetic Intermediate of Oxazole Compound and Method for Producing the Same. Otsuka Pharmaceutical Co.; 2011. [Google Scholar]
- 35.Subbian S, Tsenova L, Yang G, O’Brien P, Parsons S, Peixoto B, Taylor L, Fallows D, Kaplan G. Chronic pulmonary cavitary tuberculosis in rabbits: a failed host immune response. Open Biol. 2011;1:110016. doi: 10.1098/rsob.110016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Via LE, Lin PL, Ray SM, Carrillo J, Allen SS, Eum SY, Taylor K, Klein E, Manjunatha U, Gonzales J, Lee EG, Park SK, Raleigh JA, Cho SN, McMurray DN, Flynn JL, Barry CE., 3rd Tuberculous Granulomas are Hypoxic in Guinea pigs, Rabbits, and Non-Human Primates. Infect Immun. 2008;76:2333–2340. doi: 10.1128/IAI.01515-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waters NJ, Jones R, Williams G, Sohal B. Validation of a rapid equilibrium dialysis approach for the measurement of plasma protein binding. J Pharm Sci. 2008;97:4586–4595. doi: 10.1002/jps.21317. [DOI] [PubMed] [Google Scholar]
- 38.Di L, Umland JP, Trapa PE, Maurer TS. Impact of recovery on fraction unbound using equilibrium dialysis. J Pharm Sci. 2012;101:1327–1335. doi: 10.1002/jps.23013. [DOI] [PubMed] [Google Scholar]
- 39.Kalvass JC, Maurer TS. Influence of nonspecific brain and plasma binding on CNS exposure: implications for rational drug discovery. Biopharm Drug Dispos. 2002;23:327–338. doi: 10.1002/bdd.325. [DOI] [PubMed] [Google Scholar]
- 40.Cruciani G, Crivori P, Carrupt PA, Testa B. Molecular Fields in Quantitative Structure-Permeation Relationships: the VolSurf Approach. J Mol Struct: THEOCHEM. 2000;503:17–30. [Google Scholar]
- 41.Crivori P, Cruciani G, Carrupt PA, Testa B. Predicting blood-brain barrier permeation from three-dimensional molecular structure. J Med Chem. 2000;43:2204–2216. doi: 10.1021/jm990968+. [DOI] [PubMed] [Google Scholar]
- 42.Hall LH, Kier LB. The E-state as the basis for molecular structure space definition and structure similarity. J Chem Inf Comput Sci. 2000;40:784–791. doi: 10.1021/ci990140w. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


