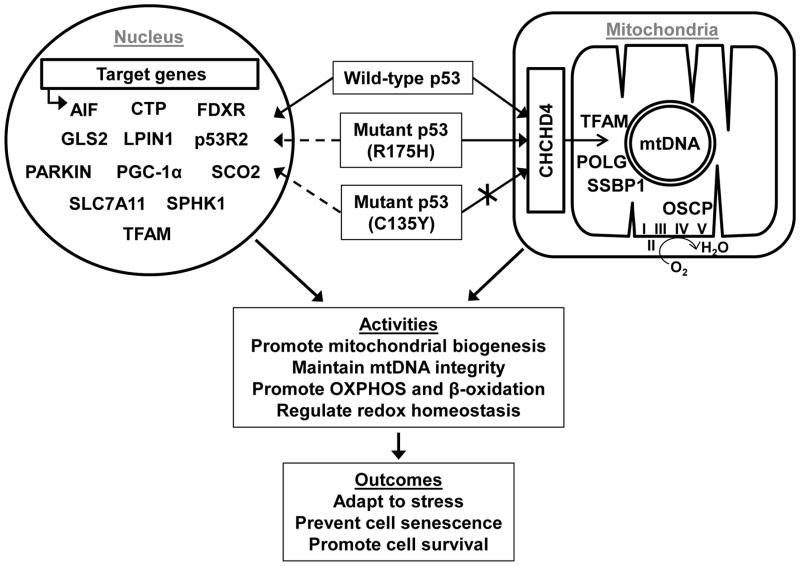
Figure 1. Transcriptional and post-translational regulation of the mitochondria by p53 is determined by multiple factors including its mutation status and translocation into the mitochondria.
Depicted are three possible genotype states: wild-type p53, mutant p53 (R175H) that can translocate into the mitochondria; and mutant p53 (C135Y) that cannot translocate into mitochondria (crossed arrow) due to disruption of its interaction with the disulfide relay protein import carrier CHCHD4. Proteins that have been reported to interact with p53 inside the mitochondria are shown along with schematic representations of circular mtDNA and respiratory complexes. p53 of all three indicated genotypes can translocate into the nucleus, but the ability to transactivate the indicated genes involved in mitochondrial function may be altered (dashed arrows) depending on its mutation status and the specific p53 target gene. Abbreviations: I, II, III, IV, and V, mitochondrial OXPHOS complexes; AIF, apoptosis-inducing factor; CTP, citrate transporter protein; FDXR, ferredoxin reductase; GLS2, mitochondrial glutaminase 2; LPIN1, lipin 1; p53R2, p53-inducible ribonucleotide reductase 2; PGC-1α, peroxisome-proliferator-activated receptor gamma co-activator-1α; SCO2, synthesis of cytochrome c oxidase 2; SLC7A11, solute carrier family 7 (cationic amino acid transporter, y+ system), member 11; SPHK1, sphingosine kinase 1; SSBP1, single-stranded DNA-binding protein; TFAM, transcription factor A, mitochondrial; OSCP, oligomycin sensitivity-conferring protein; OXPHOS, oxidative phosphorylation.