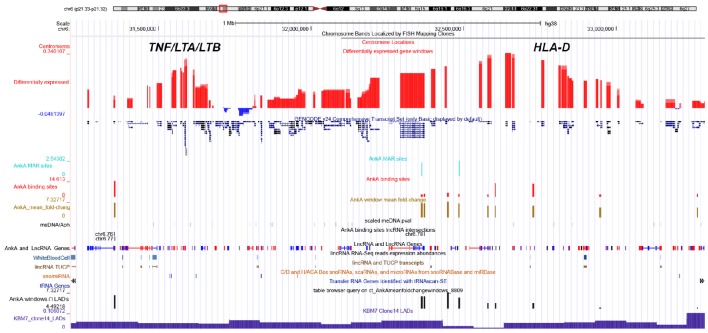
Figure 6.
UCSC genome browser comparison of genomic features and their relationship with AnkA binding at the MHC locus of chromosome 6. The top line shows the ideogram for chromosome 6 with the MHC locus identified in the red box, and the specific location is delineated by bp from the telomeric p-arm of the chromosome followed by the chromosome band and centromere location. The large red and blue bars demonstrate the average differential transcription over approximately 10 Mbp windows in human neutrophils infected ex vivo by A. phagocytophilum for 24 h, and the GENCODE v22 gene locations are shown below. Custom features are as follows from top to bottom: light blue, AnkA MAR-binding sites (height is proportional to fold change); red, AnkA binding sites converged from all three samples sets (height is proportional to fold change); gold, mean AnkA fold change over ~50 Mbp windows, centered on the midpoint of the window (height is proportional to the average fold change centered at that window); black, unique DNA methylation marks with A. phagocytophilum infection of ex vivo human neutrophils at 24 h (intensity is proportional to p-values of methylated DNA at the site); AnkA binding sites lincRNA intersections black bands—represents the intersection of AnkA binding sites with the NONCODE positions for lincRNAs and lincRNA genes; LncRNA and LncRNA Genes blue and red bands—positions and direction (red forward, blue reverse) from NONCODE; lincRNA RNA-seq reads expression abundances light blue bands—lincRNA RNA-Seq reads expression abundances for human white blood cells; lincRNA TUCP, sno/miRNA, and tRNA Genes—tracks that show locations and directions for several classes of RNA sequences that do not code for a protein; mean AnkA fold change over ~50 Mbp windows intersected with KBM7 Clone 14 LADs black bars—height of bars shows relative enrichment of AnkA at nuclear lamina; KBM7 Clone 14 LADs dark blue histogram bars—shows regions often associated with three dimensional chromatin structure that interact with the nuclear lamina and are often inactive when in this conformation. AnkA data is cumulative of all 3 donor neutrophil DNA-AnkA interactions.