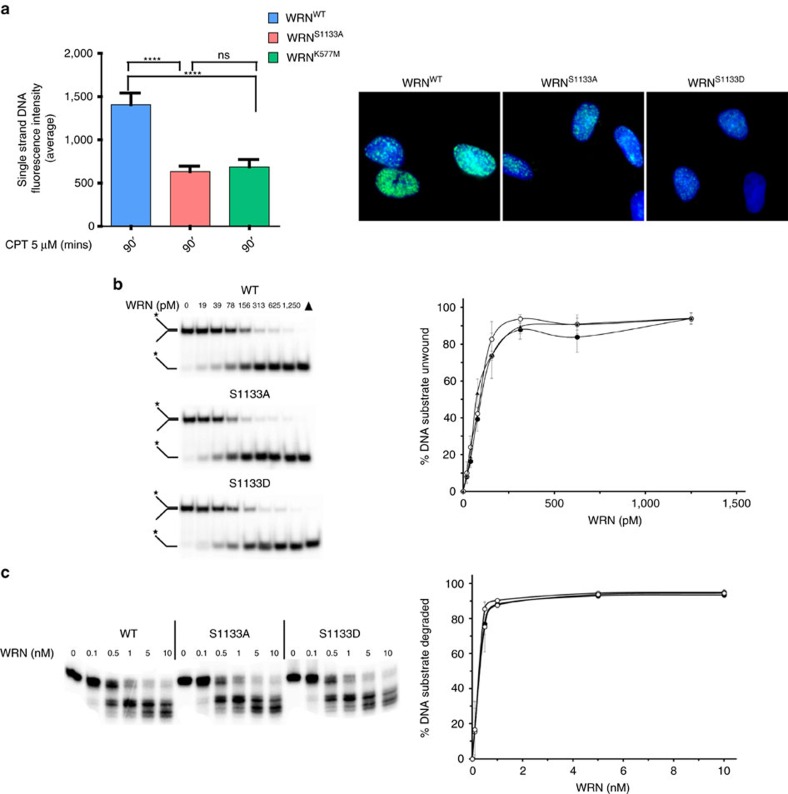
Figure 3. CDK1-dependent phosphorylation of WRN at S1133 affects the WRN helicase-DNA2-dependent pathway of end resection.
(a) WS-derived cells stably expressing the WRN wild type (WRNWT), the unphosphorylable (WRNS1133A) or the helicase-dead (WRNK577M) mutant were labelled and treated as in Fig. 2b, and end resection analysed by the non-denaturing IdU/ssDNA assay. The graph shows the mean intensity of IdU/ssDNA staining. The intensity of the anti-IdU immunofluorescence was measured from two independent experiments (n=100, each biological replicate), data are presented as mean±s.e.m. The panel shows representative images of IdU/ssDNA detection after CPT treatment. (b) Flag-WRNWT, Flag-WRNS1133A and Flag-WRNS1133D have comparable helicase activity on a forked DNA substrate. A unit of 0–1,250 pM Flag-WRNWT, Flag-WRNS1133A and Flag-WRNS1133D were incubated with forked DNA substrate as described in the Methods section. Quantitation of panels. Data represents the average of a minimum of three independent experiments. (c) Flag-WRNWT, Flag-WRNS1133A and Flag-WRNS1133D have comparable exonuclease activity on a forked DNA substrate. A unit of 0–10 nM Flag-WRNWT, Flag-WRNS1133A and Flag-WRNS1133D were incubated with forked DNA substrate as described in the Methods section. Quantitation of a. Data represent the average of a minimum of three independent experiments.