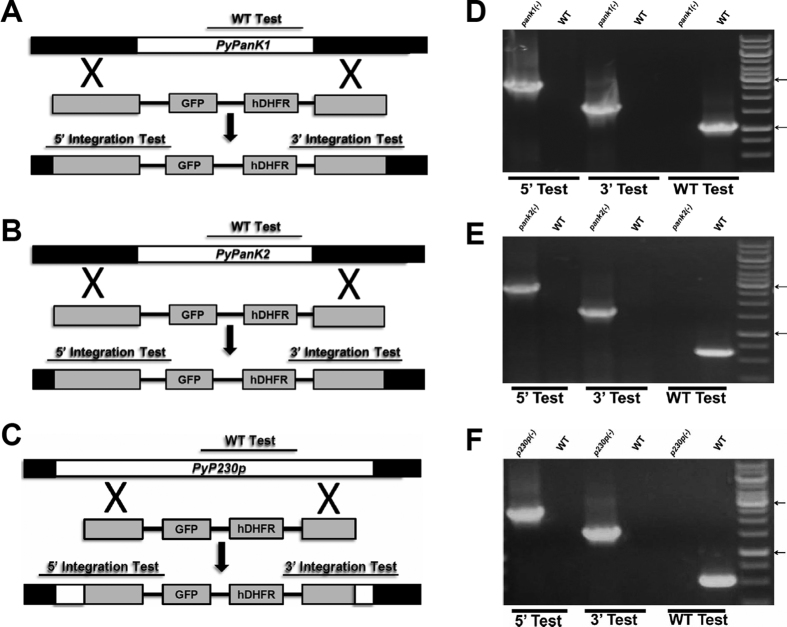
Figure 4. Targeted deletion of PyPanK1 and PyPanK2 genes.
Schematic representation of the replacement strategy to generate Pypank1(−) parasites in (A), Pypank2(−) parasites in (B) and Pyp230p(−) parasites in (C). The endogenous PyPanK1, PyPanK2 and PyP230p genomic loci are targeted with replacement fragments containing the 5′ and 3′ PyPanK1 and PyPanK2 UTRs and PyP230p ORFs sequences, respectively, flanking the human DHFR positive selection marker and eGFP cassettes. The three constructs were transfected during the same experiment, where P230p serves as a transfection experiment and transfection vector control. Diagnostic WT-specific or integration-specific test amplicons are indicated by lines. 36-cycles PCR genotyping confirms the integration of gene-replacement construct using oligonucleotide primer combinations that can only amplify from the recombinant loci (5′ Test and 3′ Test). The WT-specific PCR reaction (WT) confirms the absence of WT parasites in Pypank1(−) in (D), in Pypank2(−) parasites in (E) and in Pyp230p(−) parasites in (F).The arrows show the size of DNA ladder bands of 3000 and 1000 bps, respectively.