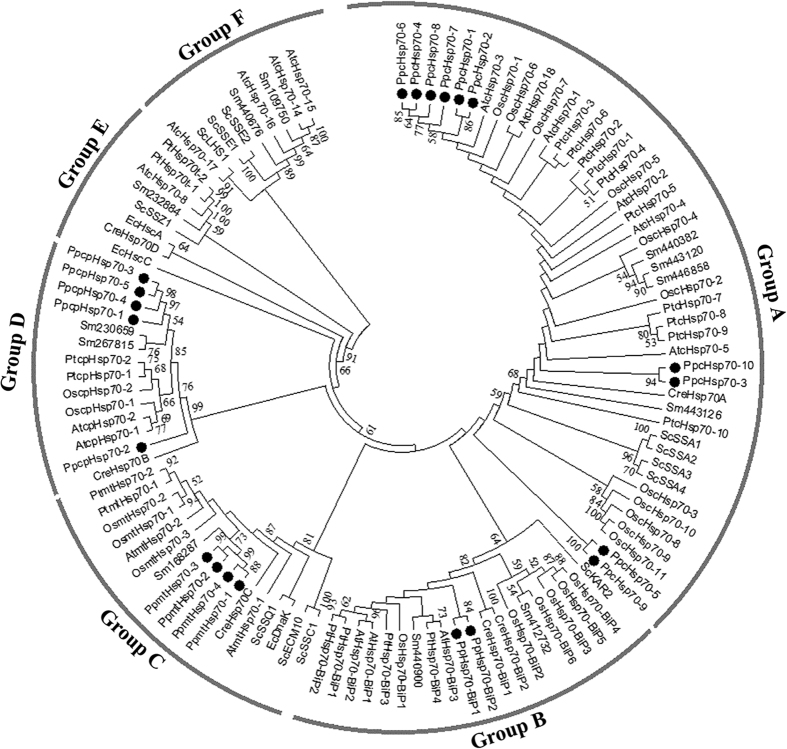
Figure 1. Phylogenetic tree of Hsp70 superfamily in eight species.
The tree was constructed using the Neighbor-Joining (NJ) method based on the amino acid sequences of Hsp70 members from Escherichia coli (Ec), Saccharomyces cerevisiae (Sc), Chlamydomonas reinhardtii (Cr), Physcomitrella patens (Pp), Selaginella moellendorffii (Sm), Oryza sativa (Os), Arabidopsis thaliana (At), and Populus trichocarpa (Pt). The Hsp70s were classified into six groups, Group A localized in the cytoplasm, Group B localized in the ER (endoplasmic reticulum), Group C localized in the mitochondrion, Group D localized in the chloroplast according to the phylogenetic analyses, Group E comprised truncated genes, and Group F was a Hsp110/SSE subfamily. The 21 Hsp70 proteins of the P. patens were marked with black dots, and were classified into 4 groups. Numbers at each branch indicate the percentage support for the node among 1,000 bootstrap replicates.