Figure 1.
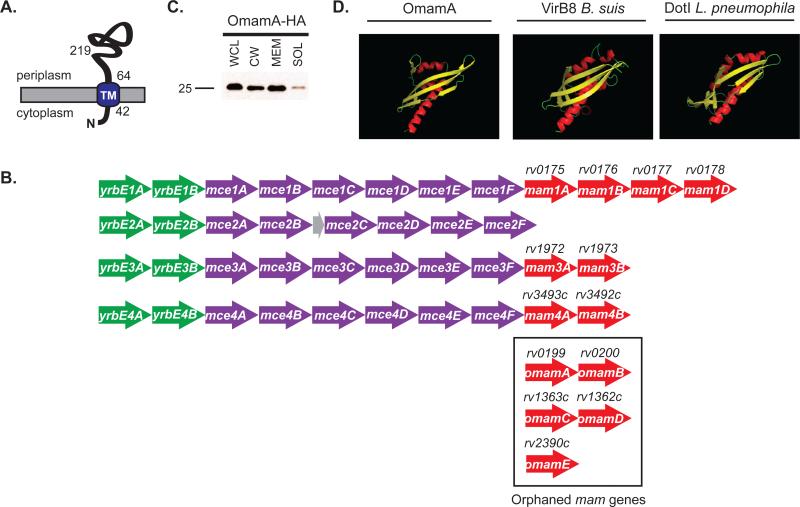
Rv0199 (OmamA) is a transmembrane protein predicted to be a Mce-associated protein. A. OmamA is predicted to have a single N-terminal transmembrane domain (TM) at amino acid 42-64 (Krogh et al., 2001) and C-terminal domain exposed to the cell wall side of the membrane (McCann et al., 2011). B. OmamA is predicted to be a Mce-associated membrane (Mam) protein, however, rv0199 is not located in a mce operon. Mce operons are typically organized by two yrbE genes upstream (green), six mce genes (purple) and most have pairs of mam genes (red) downstream. Genes encoding putative orphaned mam genes are boxed. Genes encoding Omam proteins are distinguished by being distally located from mce operons (Casali & Riley, 2007). The mce2 operon additionally contains a small predicted pseudogene (grey). C. The omamAmtb gene was engineered in frame with an HA tag and expressed in M. smegmatis. Cells were lysed to generate whole cell lysates (WCL) and fractionated by differential ultracentrifugation into cell wall (CW), cell membrane (MEM), and cytoplasmic containing soluble (SOL) fractions. Results are representative of at least three independent replicates. D. Phyre 2, an online structural prediction program, predicted with high confidence (96%) that OmamA forms a NTF2-like fold. Ribbon diagrams shown represent the Phyre 2 predicted structures of OmamA colored by secondary structure in Pymol. Ribbon diagrams representing the solved crystal structures of VirB8 from Brucella suis and DotI from Legionella pneumophila are shown for comparison. Alpha helices are colored in red, Beta-strands in yellow, and turns in green.