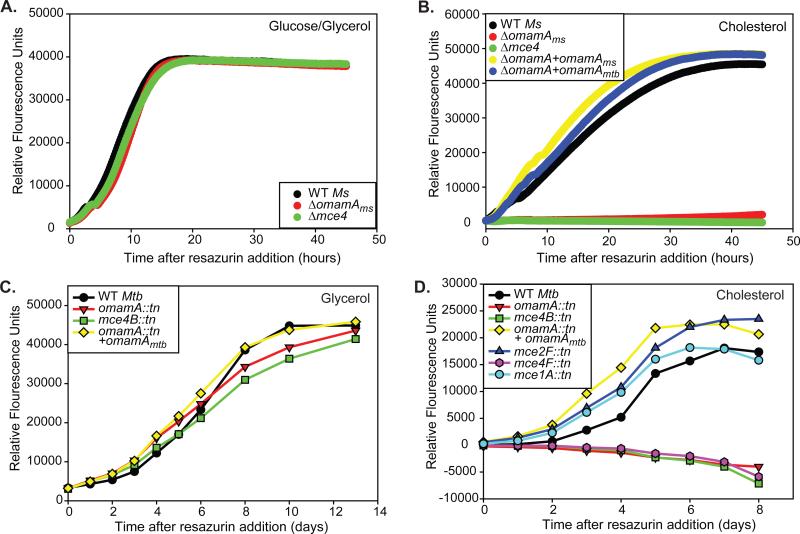
Figure 5.
OmamA is required for M. smegmatis and M. tuberculosis to utilize cholesterol. A. 104 colony forming units (cfu) of M. smegmatis strains were added to M9 glucose/glycerol and metabolic activity was monitored by resazurin conversion over time. B. 104 cfu of M. smegmatis were added to minimal M9 media plus cholesterol, and metabolic activity was monitored by resazurin conversion over time. Relative fluorescence unit measurements in cholesterol media are reported after subtraction of the minimal signal from no carbon source. 104 cfu of M. tuberculosis were added to minimal Sauton's media supplemented with C. glycerol or D. cholesterol, and metabolic activity was monitored by resazurin conversion over time. Relative fluorescence unit measurements in cholesterol media are reported after subtraction of the minimal signal from no carbon source. Results are representative of at least three independent experiments. M. smegmatis (Ms) strains: WT +pMV261 (EP1182), Δmce4 +pMV261 (EP1204), ΔomamA +pMV261 (EP1193), ΔomamA +omamAms (EP1194), and ΔomamA +omamAmtb (EP1203). M. tuberculosis (Mtb) strains: WT (MBTB178), omamA::tn (MBTB319), omamA::tn + omamA (MBTB320), mce2F::tn (MBTB156), mce1A::tn (MBTB204), mce4B::tn (MBTB329), mce4F::tn (MBTB288).