Fig. 5.
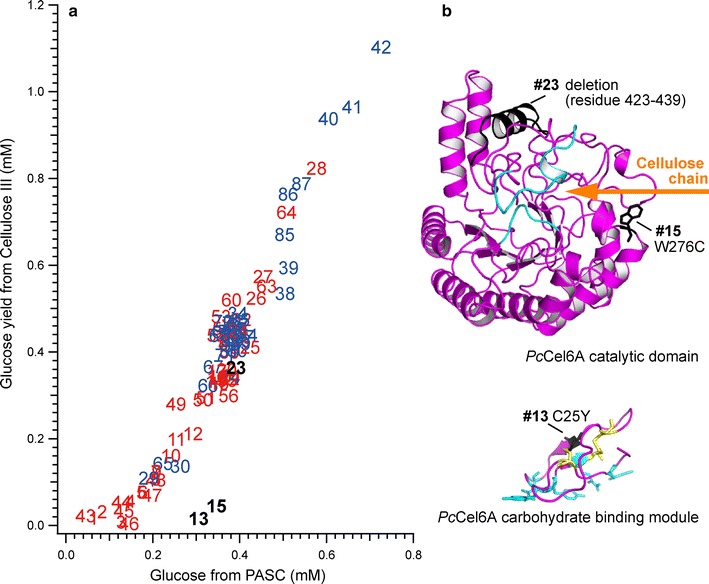
a Plot of amorphous cellulose (PASC)-degrading activity versus crystalline cellulose (cellulose IIII)-degrading activity of PcCel6A mutants. Forty-two transformants were selected from error-prone RCA-MDA plates under the conditions of 2 mM Mn2+ and 100 pg template (numbered 1–42), and 45 transformants from error-prone RCA-MDA plates under the conditions of 2 mM Mn2+ and 250 pg template (numbered 43–87), and their activities were measured. The transformants with no mutation or no change in amino acid sequence are indicated in blue. Mutants with at least one mutation in the cel6A gene are shown in red. Mutants discussed in the text are shown in black. b The mutations found in mutants #13, #15, and #23. The structures of PcCel6A catalytic domain and CBM were modeled and the locations of altered amino acids are indicated with the mutant numbers. The active site loops of the catalytic domain are colored in cyan and the direction of the incoming cellulose chain is indicated by an orange arrow. Two disulfide bridges in CBM are colored in yellow
