Abstract
We previously demonstrated that H9N2 subtype avian influenza viruses (AIVs) isolated from 1994 to 2008 evolved into distinct antigenic groups (C, D, and E) and then underwent antigenic drift from commercial vaccines, causing a country-wide outbreak during 2010–2013. In this study, H9N2 AIVs isolated from chickens during 2009–2013 were antigenically analyzed by performing hemagglutination inhibition and neutralization assays using a panel of polyclonal antibodies. Our findings confirmed the antigenic drift of recent H9N2 viruses from the commercial vaccine and showed that most of these antigenic variants form a novel HI antigenic group, F, with a few belonging to groups D and E. Slight antigenic variation was observed in group F viruses. Genetic analysis of amino acid sequences deduced from hemagglutinin (HA) gene sequences indicated that 9 of 15 mutations predominant in the 2009–2013 viruses can be mapped to known antigenic sites, which might be responsible for the novel antigenicity of group F. These antigenic changes make it necessary to modify the influenza vaccine to ensure efficient protection. A vaccine candidate, Ck/HeB/YT/10, was selected and provided significant protection against viruses from different antigenic groups in terms of reduction in virus shedding, suggesting broad cross-reactivity. Taken together, our results indicate that the H9N2 chicken influenza viruses in China have evolved from distinct antigenic groups into a novel group F that became dominant during the country-wide outbreak and now seems to be undergoing new antigenic divergence. Systematic surveillance and timely updating of vaccine strains are important for viral prevention and control in the future.
Keywords: H9N2, chicken influenza virus, antigenic evolution, vaccine
1. Introduction
H9N2 influenza virus, characterized as a low-pathogenicity pathogen, has been circulating worldwide in multiple avian species, resulting in great economic losses due to reduced egg production or high mortality associated with co-infection with other pathogens (Sun and Liu, 2015; Webster et al., 1992; Xu et al., 2007). In China, H9N2 viruses were first isolated from diseased chickens in 1994 (Chen et al., 1994); they have since become widespread in these populations (Li et al., 2005; Pu et al., 2014; Sun et al., 2010; Zhang et al., 2009). H9N2 viruses also pose a threat to public health, having caused repeated human infections in China since 1998 (Perdue and Swayne, 2005; Sun and Liu, 2015; Webster et al., 1992; Xu et al., 2007). Serologic surveillance indicates that a certain proportion (4.6%–37.2%) of people in China might be seropositive for H9N2 viruses (Liang et al., 2003; Yu et al., 2013). Since 2013, all 6 internal genes of the novel H7N9 and H10N8 viruses isolated from humans have been found in H9N2 viruses circulating in China (Chen et al., 2014; Gao et al., 2013; Pu et al., 2014). Thus, H9N2 influenza viruses are high on the list of candidates that could cause another human influenza pandemic.
To prevent H9N2 AIV infections in chickens, farms in China have administered commercially inactivated vaccines such as A/chicken/Shandong/6/1996 (Ck/SD/6/96) and A/chicken/Shanghai/F/1998 (Ck/SH/F/98) to their flocks since 1998 (Li et al., 2005; Zhang et al., 2008). However, the antigenic drift of H9N2 AIVs continues to occur and induce sporadic disease outbreaks in vaccinated chicken flocks (Huang et al., 2010; Li et al., 2005; Pu et al., 2014; Sun et al., 2012; Zhang et al., 2008; Zhang et al., 2012). Since 2010, antigenic variants of H9N2 viruses have possessed increased fitness to escape immunization pressure and caused the country-wide outbreaks in chickens that contributed to the generation of the novel H7N9 viruses (Pu et al., 2014). These facts emphasize that understanding the antigenicity of circulating viruses and updating matched vaccine strains are extremely urgent actions.
Although the antigenic drift of H9N2 chicken influenza viruses from vaccine strains was fully addressed (Li et al., 2005; Pu et al., 2014; Sun et al., 2012), the antigenic evolution of prevailing viruses isolated since 2009 is not well known. We previously investigated the H9N2 chicken influenza viruses isolated in China from 1996 to 2008 and identified 3 major antigenic groups (C, D, and E) (Sun et al., 2010). In the present study, we performed hemagglutination inhibition (HI) and neutralization assays to further investigate the antigenic evolution of the chicken H9N2 viruses isolated during 2009–2013. Additionally, we analyzed the genetic characterization of the HA gene, in which crucial substitutions might result in different antigenic properties (Boni, 2008; McHardy and Adams, 2009; Rambaut et al., 2008; Webster et al., 1992). On the basis of these data, we selected and evaluated a vaccine candidate.
2. Materials and methods
2.1. Ethics Statement
All animal research was approved by the Beijing Association for Science and Technology (approval ID SYXK, Beijing, 2007–0023) and performed in compliance with the Beijing Laboratory Animal Welfare and Ethics guide lines issued by the Beijing Administration Committee of Laboratory Animals and in accordance with the China Agricultural University (CAU) Institutional Animal Care and Use Committee guidelines (ID: SKLAB-B-2010-003) approved by the Animal Welfare Committee of CAU.
2.2. Virus
On the basis of their HA phylogenetic topology (Pu et al., 2014) and geographical distribution, 27 H9N2 chicken influenza viruses isolated from vaccinated farms from 2009 through 2013 were selected and used in this study (Fig. 1A and Table S1). These viruses were distributed across 10 provinces in China: Beijing, Guangdong, Hebei, Henan, Jiangsu, Jilin, Liaoning, Shandong, Shanxi, and Sichuan. In addition, 6 early representative viruses from identified antigenic groups (Pu et al., 2014; Sun et al., 2010) were included (Fig. 1A and Table S1). All the tested influenza viruses were sequenced in previous studies (Pu et al., 2014; Sun et al., 2010). Viruses were propagated in 10-day-old specific pathogen–free (SPF) embryonated chicken eggs. The allantoic fluid was harvested and stored at −80 °C until use.
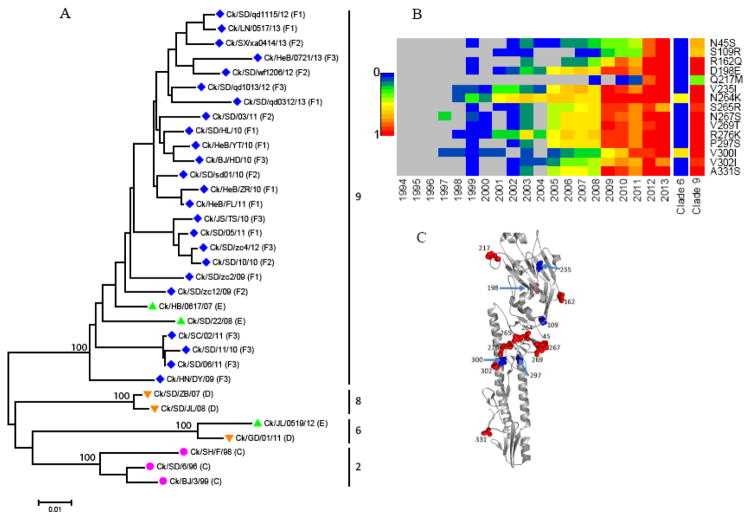
Fig. 1.
Genetic analysis of the HA genes of H9N2 chicken influenza viruses in China. (A) Phylogenetic tree of HA genes of H9N2 influenza viruses used in the HI assay. Vertical black lines mark the phylogenetic clades. Pink circles, orange inverted triangles, green triangles and blue rhombus indicate the viruses from antigenic group C, D, E, and F, respectively. Specific antigenic group names are given in parenthesis following the virus names. Abbreviations: AH, Anhui; BJ, Beijing; Ck, chicken; GD, Guangdong; HeB, Hebei; HN, Henan; JS, Jiangsu; JL, Jilin; LN, Liaoning; SD, Shandong; SH, Shanghai; SX, Shanxi; SC, Sichuan; TJ, Tianjin. (B) Prevalence of critical amino acid mutations (H9 numbering) of HA protein in H9N2 chicken viruses during 1994–2013 and in clade 6 and clade 9 viruses during 2010–2013, as compared with consensus sequences of earliest strains. Red indicates a high prevalence (up to 100%) of substitutions; blue indicates a low prevalence; and gray indicates no mutation in the given year. (C) Secondary structure of HA protein showing mutations. H9 mutations are shown as spheres with H9 numbering. Red spheres are on the surface (depth < 5 Å); blue spheres are buried.
2.3. HI assay
The HI assay was used to antigenically characterize the H9N2 viruses isolated in China from 2009 to 2013. Antisera to 10 selected H9N2 viruses were generated in 6-week-old white Leghorn SPF chickens and used in the HI assays. Among these antisera, two of them against Ck/SD/6/96 and Ck/SH/F/98 respectively were prepared in a previous study (Pu et al., 2014), and others were made in the current study. Briefly, chickens were subcutaneously vaccinated twice (at a 2-week interval) with 0.5 ml Freund’s-adjuvanted inactivated whole virus vaccines (HA content, 10 log2). Sera from vaccinated chickens were collected three weeks after the booster vaccination.
The HI test was performed using a 1% chicken red blood cell suspension as previously described (Edwards, 2006). The HI titer was expressed as the reciprocal of the highest serum dilution in which hemagglutination was inhibited.
2.4. Neutralization assay
The neutralization test was performed in SPF embryonated chicken eggs by using the diluted-serum constant-virus procedure (Lee et al., 2004). Briefly, antiserum was serially diluted 2-fold from an initial 1:10 dilution and then mixed with 100 EID50 (egg-infective dose at which 50% of inoculated eggs are infected) virus for 1 h at 37 °C. This mixture was then inoculated into five eggs. Forty-eight hours after inoculation, allantoic fluid was examined for hemagglutination activity to determine the presence of the virus. The titer was reported as the reciprocal of the highest dilution that reduced infection by at least 50% (Kobasa et al., 2004).
2.5. Antigenic cartography construction
Antigenic cartography was performed by using the program AntigenMap (http://sysbio.cvm.msstate.edu/AntigenMap), which uses matrix completion multidimensional scaling to map HI titers and neutralization titers in two dimensions (Cai et al., 2010).
2.6. Sequence collection, alignment, and phylogenetic analysis
All previously published sequences of Chinese H9N2 influenza A virus (1994–2013) were collated from FluDB (www.fludb.org), GISAID (www.gisaid.org), and NCBI (www.ncbi.nlm.nih.gov/genomes/FLU). All replicate submissions were removed by identifying sets of isolates with identical sequences. The resulting sequences were aligned by using MAFFT v6 (Katoh et al., 2002), manually adjusted to correct frame-shift errors, and subsequently translated. Downstream phylogenetic analyses were performed on the HA region of 190-1563. The unrooted phylogenetic tree was constructed by using MEGA (version 4.1) to perform the neighbor-joining method with 1000 bootstrap replicates (Zhang et al., 2009). Clade classification was based on our previous definition (Pu et al., 2014).
2.7. Vaccination and virus challenge
The A/chicken/Hebei/YT/2010 (Ck/HeB/YT/10)-containing allantoic fluid was inactivated at 37 °C for 18 h in a 0.02% nal concentration of formalin and emulsified with Freund’s adjuvant as previously described (Tumpey et al., 2001). The oil-emulsion inactivated vaccine prepared from Ck/HeB/YT/10 was used for vaccination. These experiments were designed as previously described (Pu et al., 2014) and were performed in unvaccinated and vaccinated chickens. Three-week-old SPF white Leghorn chickens were subcutaneously vaccinated twice (at a 2-week interval) with a Ck/HeB/YT/10-based vaccine (HA content, 10 log2). HI titers to homologous antigen were measured in 2-fold serum dilutions 2 weeks after the second inoculation and expressed as the log2 reciprocal of the end point. The vaccinated chickens were divided into 2 groups for challenge experiments: those with a low antibody level (HI titer, 6–9) and those with a high antibody level (HI titer, 10–12). Seven-week-old SPF white Leghorn chickens were used as the unvaccinated group. Eight H9N2 viruses were used to inoculate chickens. In inoculated groups, birds (n = 10 per group) were inoculated intranasally with 106 EID50 of each virus in a 0.2-mL volume. Tracheal and cloacal swabs were collected at 3, 5, and 7 days post inoculation (dpi). The virus titers of positive samples were measured by determining the EID50; the lower limit of detection was 1.0 log10EID50/mL. In contact groups, 5 chickens were placed in physical contact (i.e., in the same cage, sharing food and water) with inoculated birds 24 h after inoculation. Tracheal and cloacal swabs were collected at 3, 5, and 7 dpi for viral detection and titration, as just described.
2.8. Statistical Analysis
Experimental groups were statistically compared by using Prism 5.0 software (GraphPad Software, La Jolla, CA) to perform ANOVA. P < 0.05 was considered to indicate a statistically significant difference.
3. Results
3.1. Antigenic analysis of H9N2 chicken influenza viruses
Among the 27 newly identified viruses isolated in 2009–2013, 3 belonged to the previously identified antigenic groups D and E, and all others formed a novel antigenic group, F (Fig. 2 and Table S1). Most group F viruses had low or moderate reactivity to group C and E antisera but reacted well with group D and F antisera (Table S1). Further comparison of groups D and F showed that group F viruses have a different antigenic profile in reactions with antisera to the recent viruses Ck/HeB/YT/10, Ck/GD/01/11, Ck/JS/TS/10, and Ck/SD/06/11. The group C viruses Ck/SD/6/96 and Ck/SH/F/98 are the commercial vaccine strains commonly used in chickens since 1998. The low reactivity to vaccine antisera indicated that the recent H9N2 viruses are antigenically distinct from the vaccine strains, which is similar to our previous finding (Pu et al., 2014). In the described HI assay, the viruses from all identified antigenic groups showed high reactivity to the antisera of Ck/HeB/YT/10 virus. Their titers were comparable to or 2- to 8-fold higher than that of the homologous virus (Table S1).
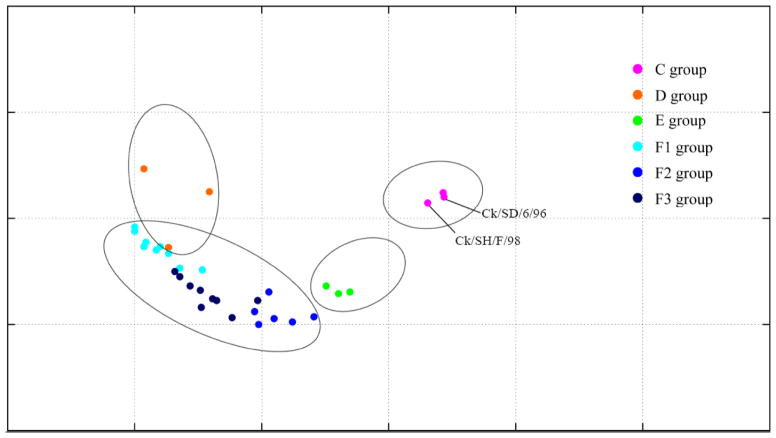
Fig. 2.
Antigenic cartography representation of the HI data generated by using a panel of chicken antisera. Details of the HI data are shown in Table S1. The map was produced by using AntigenMap (http://sysbio.cvm.msstate.edu/AntigenMap). One unit (grid) represents a 2-fold change in the HI assay results. Viruses in the same HI group are encircled in an oval.
The group F viruses had relatively different antigenicities (Fig. 2 and Table S1); therefore, we further categorized them into 3 subgroups: F1, F2, and F3. Subgroup F1 viruses had the lowest HI titers in reactions with the antisera to group C viruses and some of group F viruses (Ck/HeB/YT/10 and Ck/SD/06/11) (Table S1). Most subgroup F2 viruses reacted best with the antisera to viruses Ck/GD/01/11 and Ck/JS/TS/10, inducing HI titers that were 2 to 16 times greater than those of F1 and F3 viruses. Subgroup F3 viruses had antigenicity similar to that of F1 viruses but reacted better with the antiserum of Ck/HeB/YT/10, Ck/SD/06/11, and Ck/GD/01/11 than did F1 viruses.
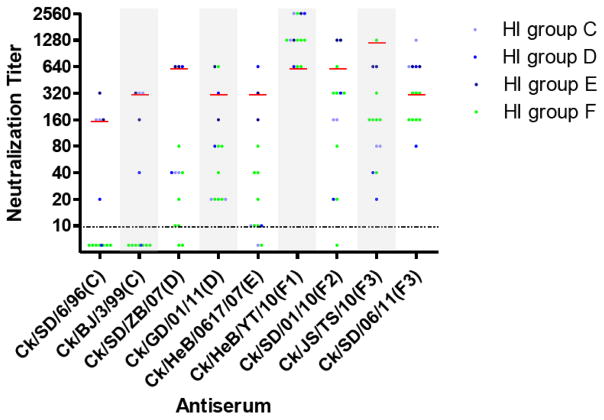
To further confirm the antigenicity of H9N2 chicken influenza viruses and determine their antisera’s neutralizing activity, we performed neutralization assays with multiple polyclonal antisera and representative viruses from each HI antigenic group. The neutralizing reaction profiles show that these recent viruses of HI group F had more-obvious antigenicity differences than did early homologous viruses of groups C, D, and E and had at least 4-fold lower neutralization titers upon reaction with antisera raised against group C, D, or E viruses (Fig. 3, Fig. S1 and Table S2). Group F viruses also showed antigenic diversity in the neutralization assay: some of them reacted weakly with the antisera against Ck/SD/sd01/10 and Ck/JS/TS/10, resulting in a 2-to 32-fold decrease in titers (Fig. 3 and Table S2). Consistent with the results of the HI assay, Ck/HeB/YT/10 antisera elicited cross-reactive neutralizing antibodies to the heterologous virus and had titers that were equal to or 2-to 4-fold higher than that of the homologous neutralizing antibody (Fig. 3 and Table S2).
Fig. 3.
Neutralization assay of H9N2 chicken influenza viruses andantisera. HI group C (n = 2), group D (n = 2), and group F (n = 7) viruses were selected for the neutralization assays and used with antisera raised against viruses from different HI groups. Details of viruses and antisera are shown in Table S2. The dashed line indicates the lowest limit of detection (Neutralization titer = 10). The red lines indicate homologous titers.
Combining the results of HI and neutralization assays shows that the newly identified HI group F exhibited antigenicity that is distinct from that of groups C, D, and E. Among the group F viruses, slight antigenic variation was found, indicating the antigenic diversity of H9N2 chicken influenza viruses circulating in China. Ck/HeB/YT/10 consistently had the best cross-reactive response to all of the prevalent H9N2 viruses and was selected as an ideal vaccine candidate.
3.2. Molecular characteristics of HA genes of H9N2 chicken influenza viruses
Antigenic change is due mainly to point mutations in the HA gene of influenza viruses (Boni, 2008; McHardy and Adams, 2009; Rambaut et al., 2008; Webster et al., 1992). To investigate the molecular characteristics of the HA genes of H9N2 influenza viruses in China, the deduced amino acid sequences were aligned by using all available HA sequences from 1994 through 2013. Fifteen mutations were identified as predominant in the 2009–2013 viruses (Fig. 1B). Phylogenetically, recent viruses belong mainly to clades 6 and 9 in the HA tree (Fig. 1A). Further analysis showed that these amino acid mutations in recent viruses are predominant in the prevalent clade 9 viruses (represented by the group F virus Ck/HeB/YT/10/11) but not in clade 6 viruses (represented by the group D virus Ck/GD/01/11) (Fig. 1B).
These substitutions were mainly on the surface of the HA1 globular head (Fig. 1C). Nine of them can be mapped to the known antigenic sites of H1 (Caton et al., 1982), H2 (Tsuchiya et al., 2001), H3 (Wiley et al., 1981), H5 (Zou et al., 2012), and H9 (Kaverin et al., 2004) serotypes (Table S3). Another HA mutation, P297S, introduces a new glycosylation site at Asn295, consistent with a strategy commonly used by influenza viruses to mask and unmask antigenic sites from the immune system (Baigent and McCauley, 2001; Perdue and Suarez, 2000). Therefore, the distinct antigenicity of HI group F from that of other groups may have been induced by these mutants, which have been predominant since 2009.
Specific amino acid mutations were also observed in the representative isolates from different subgroups of group F. For example, subgroup F2 virus Ck/JS/TS/11 has the N45G mutation in an antigenic site of H5N1 viruses, which might cause antigenic diversification of these subgroups (data not shown).
3.3. Protective efficacy of Ck/HeB/YT/10 vaccine candidate in chickens
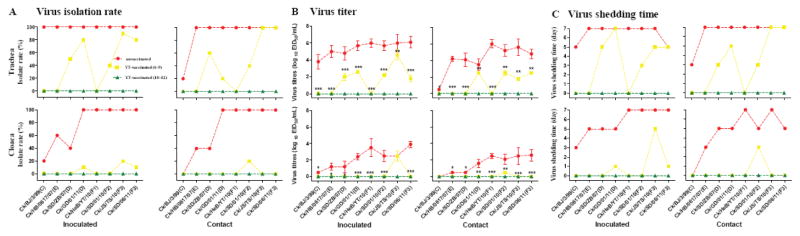
Because Ck/HeB/YT/10 elicited good cross-reactions, we selected it as a vaccine candidate. In the unvaccinated groups, all of the tested viruses consistently showed efficient replication in tracheas of inoculated chickens, with titers of 3.8 to 6.1 EID50/mL and a 100% isolation rate (Fig. 4A and Fig. 4B). These tested viruses were also shed from the cloaca, with a detection rate of 20% to 100% (Fig. 4A). Most viruses maintained their growth in trachea and cloaca as long as 7 days (Fig. 4C). All of the test viruses transmitted efficiently (100%) except for Ck/BJ/3/99 (Fig. 4A).
Fig. 4.
Protective efficacy of vaccine candidate Ck/HeB/YT/10 in chickens. Freund’s adjuvanted inactivated vaccine based on Ck/HeB/YT/10 was used for vaccination. Protective efficacy was evaluated by determining the viral replication and transmission in vaccinated chickens with low antibody titers of 6–9 (yellow dashed line) and in vaccinated chickens with high titers of 10–12 (green dashed line). Unvaccinated chickens were used as controls (red dashed line). The dashed lines show the differences of infection among the viruses from the different antigenic groups. Eight representative H9N2 chicken viruses selected from HI groups C, D, E, or F were tested to determine their infectivity in inoculated and contact chickens. (A) Isolation rates of H9N2viruses in chickens. (B) Mean titers of H9N2 viruses at 3 dpi. Error bars represent SD. The titers in vaccinated chickens (with antibody titers of 6–9) were significantly lower than those in unvaccinated chickens (*P < 0.05, **P < 0.01,***P < 0.001; one-way ANOVA). (C) The viral shedding time of H9N2 viruses.
Upon challenge in the groups with low antibody levels, immunization with Freund’s adjuvanted inactivated vaccine Ck/HeB/YT/10 provided complete protection against virus shedding of group C, group E, and homologous (Ck/HeB/YT/10) viruses in trachea and cloaca (Fig. 4A) and partial protection against other viruses, as shown by a 10% to 80% lower isolation rate in trachea and an 80% to 90% lower isolation rate in cloaca (Fig. 4A). Consistently, the viral titers significantly decreased at 3 dpi (P < 0.05, ANOVA) (Fig. 4B). Most viruses were shed for a shorter time: they were recovered from trachea for 3–5 days and from cloaca for 1–3 days (Fig. 4C). However, in the high antibody level groups, vaccination elicited increased protection efficacy, and none of the tested viruses were detected in both inoculated and contact groups (Fig. 4A to 4C). In summary, these results demonstrated that vaccination with vaccine Ck/HeB/YT/10 efficiently inhibited the replication and transmission of H9N2 AIVs from different antigenic groups, especially in vaccinated chickens with high antibody levels.
4. Discussion
H9N2 viruses isolated during a certain period or in a certain region were previously believed to share similar antigenicity profiles (Webster et al., 1992; Xu et al., 2007; Zhang et al., 2008). Therefore, vaccine strains isolated during the late 20th century were used in chicken flocks throughout China for more than 10 years (Li et al., 2005; Sun et al., 2012). However, our and other previous studies revealed that some H9N2 chicken influenza viruses isolated especially since 2007 underwent antigenic drift from the vaccine strains and formed distinct antigenic groups (Li et al., 2005; Pu et al., 2014; Sun et al., 2010; Zhang et al., 2012). In 2010–2013, antigenic variants finally caused a country-wide outbreak of H9N2 viruses in chickens in China (Pu et al., 2014).
In a previous study, we found that H9N2 isolates from 2009 to 2011 underwent significant antigenic drift from the vaccine strains (Ck/SD/6/96 and Ck/SH/F/98) (Pu et al., 2014). In this study, we confirmed the antigenic drift of 2009–2011 viruses and demonstrated the antigenic changes also in 2012–2013 viruses, and further showed that the 2009–2013 variants exhibited different antigenic profiles from group C, D, and E viruses, forming a novel HI antigenic group, F. Phylogenetically, viruses from the previous distinct antigenic groups C, D, and E, which were mainly isolated before 2009, were in clades 2, 6, 8, and 9 in the HA tree; but, the antigenic group F variants were all in clade 9. The predominant antigenic mutations present in the clade 9 viruses of 2009–2013 further indicated that group F viruses are genetically different from the viruses belonging to groups C, D, and E. At the genotype level, H9N2 viruses of groups C, D, and E were previously demonstrated to belong to multiple genotypes, but these current variants were mostly from genotype G57 of H9N2 viruses, which had increased dramatically since 2009 and became predominant during 2010–2013 (Pu et al., 2014). Therefore, group F viruses shared genetic and antigenic characteristics different from viruses of groups C, D, and E; they evolved from distinct antigenic groups, caused the country-wide outbreak, and became dominant during the period. Genetic differences, especially those in HA gene, may be responsible for the antigenic divergence of group F from previous virus groups.
Since the country-wide outbreak of H9N2 viruses in chickens, this escape from immunological pressure may have contributed to further antigenic variation among viruses. Clades 6 and 9 were the 2 major circulating lineages of H9N2 viruses in 2011, but the representative viruses from these 2 clades respectively belong to HI groups D and F, which have distinct antigenicity. Group F viruses also have relatively different antigenicities from each other and can be categorized into 3 subgroups: F1, F2, and F3. Different subgroups of viruses were detected within a given geographic region or at a given time point. For example, H9N2 viruses of F1, F2, and F3 were co-circulating in Shandong Province during 2012–2013.
Viruses sharing a close genetic relationship also diverged into different antigenic groups. For instance, Ck/GD/01/11 and Ck/JL/0519/12 both belong to clade 6 but fall into antigenic groups D and E, respectively. This discrepancy may be due to the relationship between antigenic change and genetic change being nonlinear below the clade level (McHardy and Adams, 2009). Some mutations may exert a disproportionately large effect on the antigenic type, whereas others are “hitchhikers” with no phenotypic effect (McHardy and Adams, 2009). The described results suggest that after escaping, H9N2 viruses circulating in China are transiting from a stage of decreased antigenic diversity to a stage of increased diversity. The antigenic diversity of prevailing H9N2 viruses highlights the importance of selecting a vaccine candidate that is a good antigenic match with prevailing strains and is broadly cross-reactive.
Ck/HeB/YT/10 virus was selected here as a vaccination candidate because it was from the prevalent clade 9 and was highly cross-reactive with homologous and heterologous viruses in the HI and neutralization assays. As expected, vaccination with Freund’s adjuvanted inactivated vaccine Ck/HeB/YT/10 efficiently inhibited the shedding of H9N2 AIVs from different antigenic groups in vaccinated chickens with high-antibody levels, unlike the A/chicken/Shandong/6/1996-based commercial vaccine, which failed to provide protection against these prevailing viruses (Pu et al., 2014). This protection difference can be explained by the data from the HI and neutralization assay. The antiserum to Ck/SD/6/96 virus from HI antigenic group C consistently showed low reactivity with the prevailing viruses especially from group D and E; however, the antiserum against Ck/HeB/YT/10 had strong cross reactions with all viruses of group C, D and E. These results suggest that Ck/HeB/YT/10 can be used as an ideal broad-spectrum vaccine strain that may provide efficient protection against the prevailing H9N2 viruses from different regions.
Partial protection was observed in some challenged chickens with low antibody titers, which is common in the development of vaccines (Bertran et al., 2013; Park et al., 2014; Tian et al., 2005). In the field, the sera antibody titers of farmed chickens are generally 10 to 12 log2 after vaccination with multiple doses (Cai et al., 2009; Tong et al., 2010). Therefore, in using the candidate vaccine strain, it is necessary to maintain high antibody titers in chicken flocks. In addition, veterinary biosecurity measures, including strict administration of live-poultry markets, segregation, cleaning, and disinfection, should be highly intensified to prevent and control avian influenza.
Conclusion
Taken together, our findings demonstrate the newly antigenic changes of H9N2 chicken influenza viruses in China from 2009–2013. The tendency of the novel group F viruses toward antigenic diversity highlights the necessity of monitoring antigenic variants in chickens. Vaccine strains should be systematically evaluated and regularly updated to achieve optimal protection against AIV infections.
Supplementary Material
Highlights.
2009–2013 H9N2 chicken viruses evolved into a novel antigenic group F in China.
Group F viruses are responsible for the recent country-wide outbreak of H9N2.
Slight antigenic variation in group F indicates an antigenic divergence trend.
A newly evaluated H9N2 vaccine candidate inhibited the viral infection in chickens.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 3143008 and 2015DY004); the National High Technology Research and Development Program (863 Program) of China (Grant No.2012AA101303); the China Agriculture Research System; and the Poultry-Related Science and Technology Innovation Team of Peking. This work was also funded in part by Contract numbers HHSN266200700005C and HHSN272201400006C from the National Institute of Allergy and Infectious Disease, National Institutes of Health, Department of Health and Human Services, and by the American Lebanese Syrian Associated Charities (ALSAC).
Footnotes
Conflicts of interest: None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baigent SJ, McCauley JW. Glycosylation of haemagglutinin and stalk-length of neuraminidase combine to regulate the growth of avian influenza viruses in tissue culture. Virus research. 2001;79:177–185. doi: 10.1016/s0168-1702(01)00272-6. [DOI] [PubMed] [Google Scholar]
- Bertran K, Sá e Silva M, Pantin-Jackwood MJ, Swayne DE. Protection against H7N3 high pathogenicity avian influenza in chickens immunized with a recombinant fowlpox and an inactivated avian influenza vaccines. Vaccine. 2013;31:3572–3576. doi: 10.1016/j.vaccine.2013.05.039. [DOI] [PubMed] [Google Scholar]
- Boni MF. Vaccination and antigenic drift in influenza. Vaccine. 2008;26:C8–C14. doi: 10.1016/j.vaccine.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Qi WB, Xu CG, Chen XC, Luo KJ, Xiao ZY, Zhang GH, Liao M. Immunization program of the inactivated bivalent influenza vaccine (H5+H9)in breeder chickens. Chinese Journal of Veterinary Medicine. 2009;45:14–16. [Google Scholar]
- Cai Z, Zhang T, Wan XF. A computational framework for influenza antigenic cartography. PLoS computational biology. 2010;6:e1000949. doi: 10.1371/journal.pcbi.1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype) Cell. 1982;31:417–427. doi: 10.1016/0092-8674(82)90135-0. [DOI] [PubMed] [Google Scholar]
- Chen B, Zhang Z, Chen W. The study of avian influenza: I. The isolation and preliminary serological identification of avian influenza virus in chicken. Chinese Journal of Veterinary Medicine. 1994:3–5. [Google Scholar]
- Chen H, Yuan H, Gao R, Zhang J, Wang D, Xiong Y, Fan G, Yang F, Li X, Zhou J. Clinical and epidemiological characteristics of a fatal case of avian influenza A H10N8 virus infection: a descriptive study. The Lancet. 2014;383:714–721. doi: 10.1016/S0140-6736(14)60111-2. [DOI] [PubMed] [Google Scholar]
- Edwards S. OIE laboratory standards for avian influenza. Developments in biologicals. 2006;124:159–162. [PubMed] [Google Scholar]
- Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, Xu X, Lu H, Zhu W, Gao Z, Xiang N, Shen Y, He Z, Gu Y, Zhang Z, Yang Y, Zhao X, Zhou L, Li X, Zou S, Zhang Y, Li X, Yang L, Guo J, Dong J, Li Q, Dong L, Zhu Y, Bai T, Wang S, Hao P, Yang W, Zhang Y, Han J, Yu H, Li D, Gao GF, Wu G, Wang Y, Yuan Z, Shu Y. Human infection with a novel avian-origin influenza A (H7N9) virus. The New England journal of medicine. 2013;368:1888–1897. doi: 10.1056/NEJMoa1304459. [DOI] [PubMed] [Google Scholar]
- Huang Y, Hu B, Wen X, Cao S, Gavrilov BK, Du Q, Khan MI, Zhang X. Diversified reassortant H9N2 avian influenza viruses in chicken flocks in northern and eastern China. Virus Res. 2010;151:26–32. doi: 10.1016/j.virusres.2010.03.010. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaverin NV, Rudneva IA, Ilyushina NA, Lipatov AS, Krauss S, Webster RG. Structural differences among hemagglutinins of influenza A virus subtypes are reflected in their antigenic architecture: analysis of H9 escape mutants. Journal of virology. 2004;78:240–249. doi: 10.1128/JVI.78.1.240-249.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobasa D, Takada A, Shinya K, Hatta M, Halfmann P, Theriault S, Suzuki H, Nishimura H, Mitamura K, Sugaya N. Enhanced virulence of influenza A viruses with the haemagglutinin of the 1918 pandemic virus. Nature. 2004;431:703–707. doi: 10.1038/nature02951. [DOI] [PubMed] [Google Scholar]
- Lee CW, Senne DA, Suarez DL. Effect of vaccine use in the evolution of Mexican lineage H5N2 avian influenza virus. J Virol. 2004;78:8372–8381. doi: 10.1128/JVI.78.15.8372-8381.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Yu K, Tian G, Yu D, Liu L, Jing B, Ping J, Chen H. Evolution of H9N2 influenza viruses from domestic poultry in Mainland China. Virology. 2005;340:70–83. doi: 10.1016/j.virol.2005.06.025. [DOI] [PubMed] [Google Scholar]
- Liang Q, Li J, Chen P, Chen Y, Xuan W, Ning J, Wei Y, Li K. Seroepidemiologic survey for antibody of healthy youth to H9, H6 and H5 subtypes of influenza A virus in Chaoshan Area. J Shantou University Medical College. 2003;16:107–108. [Google Scholar]
- McHardy AC, Adams B. The role of genomics in tracking the evolution of influenza A virus. PLoS pathogens. 2009;5:e1000566. doi: 10.1371/journal.ppat.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JK, Lee DH, Cho CH, Yuk SS, To EO, Kwon JH, Noh JY, Kim BY, Choi SW, Shim BS, Song MK, Lee JB, Park SY, Choi IS, Song CS. Supplementation of oil-based inactivated H9N2 vaccine with M2e antigen enhances resistance against heterologous H9N2 avian influenza virus infection. Veterinary Microbiology. 2014;169:211–217. doi: 10.1016/j.vetmic.2014.01.001. [DOI] [PubMed] [Google Scholar]
- Perdue ML, Suarez DL. Structural features of the avian influenza virus hemagglutinin that influence virulence. Veterinary microbiology. 2000;74:77–86. doi: 10.1016/s0378-1135(00)00168-1. [DOI] [PubMed] [Google Scholar]
- Perdue ML, Swayne DE. Public health risk from avian influenza viruses. Avian Dis. 2005;49:317–327. doi: 10.1637/7390-060305R.1. [DOI] [PubMed] [Google Scholar]
- Pu J, Wang S, Yin Y, Zhang G, Carter RA, Wang J, Xu G, Sun H, Wang M, Wen C. Evolution of the H9N2 influenza genotype that facilitated the genesis of the novel H7N9 virus. Proceedings of the National Academy of Sciences; 2014. 201422456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Pybus OG, Nelson MI, Viboud C, Taubenberger JK, Holmes EC. The genomic and epidemiological dynamics of human influenza A virus. Nature. 2008;453:615–619. doi: 10.1038/nature06945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Liu J. H9N2 influenza virus in China: a cause of concern. Protein & cell. 2015;6:18–25. doi: 10.1007/s13238-014-0111-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Pu J, Fan L, Sun H, Wang J, Zhang Y, Liu L, Liu J. Evaluation of the protective efficacy of a commercial vaccine against different antigenic groups of H9N2 influenza viruses in chickens. Veterinary microbiology. 2012;156:193–199. doi: 10.1016/j.vetmic.2011.10.003. [DOI] [PubMed] [Google Scholar]
- Sun Y, Pu J, Jiang Z, Guan T, Xia Y, Xu Q, Liu L, Ma B, Tian F, Brown E. Genotypic evolution and antigenic drift of H9N2 influenza viruses in China from 1994 to 2008. Veterinary microbiology. 2010;146:215–225. doi: 10.1016/j.vetmic.2010.05.010. [DOI] [PubMed] [Google Scholar]
- Tian G, Zhang S, Li Y, Bu Z, Liu P, Zhou J, Li C, Shi J, Yu K, Chen H. Protective efficacy in chickens, geese and ducks of an H5N1-inactivated vaccine developed by reverse genetics. Virology. 2005;341:153–162. doi: 10.1016/j.virol.2005.07.011. [DOI] [PubMed] [Google Scholar]
- Tong HB, Zhang XR, Wu YT, Liu XX, Gong JS, Liu XF. Phylogenetic analysis of H9N2 subtype avian influenza viruses isolated from immunized chicken flocks and protective efficacy of F strain inactivated vaccine. Chinese Journal of Preventive Veterinary Medicine. 2010;32:933–936. [Google Scholar]
- Tsuchiya E, Sugawara K, Hongo S, Matsuzaki Y, Muraki Y, Li ZN, Nakamura K. Antigenic structure of the haemagglutinin of human influenza A/H2N2 virus. Journal of General Virology. 2001;82:2475–2484. doi: 10.1099/0022-1317-82-10-2475. [DOI] [PubMed] [Google Scholar]
- Tumpey TM, Renshaw M, Clements JD, Katz JM. Mucosal delivery of inactivated influenza vaccine induces B-cell-dependent heterosubtypic cross-protection against lethal influenza A H5N1 virus infection. J Virol. 2001;75:5141–5150. doi: 10.1128/JVI.75.11.5141-5150.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiological reviews. 1992;56:152–179. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley D, Wilson I, Skehel J. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature. 1981;289:373–378. doi: 10.1038/289373a0. [DOI] [PubMed] [Google Scholar]
- Xu KM, Smith GJ, Bahl J, Duan L, Tai H, Vijaykrishna D, Wang J, Zhang JX, Li KS, Fan XH, Webster RG, Chen H, Peiris JS, Guan Y. The genesis and evolution of H9N2 influenza viruses in poultry from southern China, 2000 to 2005. J Virol. 2007;81:10389–10401. doi: 10.1128/JVI.00979-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Q, Liu L, Pu J, Zhao J, Sun Y, Shen G, Wei H, Zhu J, Zheng R, Xiong D, Liu X, Liu J. Risk perceptions for avian influenza virus infection among poultry workers, China. Emerging infectious diseases. 2013;19:313–316. doi: 10.3201/eid1902.120251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Tang Y, Liu X, Liu W, Zhang X, Liu H, Peng D, Gao S, Wu Y, Zhang L, Lu S, Liu X. A novel genotype H9N2 influenza virus possessing human H5N1 internal genomes has been circulating in poultry in eastern China since 1998. J Virol. 2009;83:8428–8438. doi: 10.1128/JVI.00659-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Tang Y, Liu X, Peng D, Liu W, Liu H, Lu S, Liu X. Characterization of H9N2 influenza viruses isolated from vaccinated flocks in an integrated broiler chicken operation in eastern China during a 5 year period (1998–2002) The Journal of general virology. 2008;89:3102–3112. doi: 10.1099/vir.0.2008/005652-0. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yin Y, Bi Y, Wang S, Xu S, Wang J, Zhou S, Sun T, Yoon KJ. Molecular and antigenic characterization of H9N2 avian influenza virus isolates from chicken flocks between 1998 and 2007 in China. Vet Microbiol. 2012;156:285–293. doi: 10.1016/j.vetmic.2011.11.014. [DOI] [PubMed] [Google Scholar]
- Zou W, Ke J, Zhu J, Zhou H, Jin M. The antigenic property of the H5N1 avian influenza viruses isolated in central China. Virol J. 2012;9:148. doi: 10.1186/1743-422X-9-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.