Abstract
Marine-to-terrestrial transition represents one of the most fundamental shifts in microbial life. Understanding the distribution and drivers of soil microbial communities across coastal ecosystems is critical given the roles of microbes in soil biogeochemistry and their multifaceted influence on landscape succession. Here, we studied the fungal community dynamics in a well-established salt marsh chronosequence that spans over a century of ecosystem development. We focussed on providing high-resolution assessments of community composition, diversity and ecophysiological shifts that yielded patterns of ecological succession through soil formation. Notably, despite containing 10- to 100-fold lower fungal internal transcribed spacer abundances, early-successional sites revealed fungal richnesses comparable to those of more mature soils. These newly formed sites also exhibited significant temporal variations in β-diversity that may be attributed to the highly dynamic nature of the system imposed by the tidal regime. The fungal community compositions and ecophysiological assignments changed substantially along the successional gradient, revealing a clear signature of ecological replacement and gradually transforming the environment from a marine into a terrestrial system. Moreover, distance-based linear modelling revealed soil physical structure and organic matter to be the best predictors of the shifts in fungal β-diversity along the chronosequence. Taken together, our study lays the basis for a better understanding of the spatiotemporally determined fungal community dynamics in salt marshes and highlights their ecophysiological traits and adaptation in an evolving ecosystem.
Introduction
Salt marshes are highly productive intertidal wetlands located in temperate climatic zones. In these ecosystems, plant litter decomposition is impaired by the low temperatures and low litter quality, combined with salinity and the dynamic switches between oxic and anoxic conditions. Collectively, these factors lead to elevated levels of carbon (C) sequestration and long-term organic matter storage, which in salt marshes occurs at rates that outpace many other temperate biomes (Chmura et al., 2003). In many terrestrial ecosystems, fungi act as major players in the C cycle by influencing C sequestration—either positively or negatively (Heinemeyer et al., 2007)—and mediating the allocation of a major fraction of photosynthetically fixed C from plants into soil (Högberg et al., 2001; Heimann and Reichstein, 2008). In salt marshes, a few studies have partitioned the role of arbuscular mycorrhizal (AM) fungi that associate with endemic plant species (Hildebrandt et al., 2001; Wilde et al., 2009). However, studies on the composition and distribution of fungal communities in soil have thus far only been conducted over relatively small scales. Moreover, these investigations are often based on approaches that lack taxonomic resolution or considerations of spatial and temporal variations (Burke et al., 2002; Buchan et al., 2002; Torzilli et al., 2006). Hence, we still understand relatively little of how fungal communities establish in salt marshes and the degree to which different (biotic and abiotic) factors drive taxon distributions. These have a direct influence in the rate of community turnover, which implicates in shifting ecological roles of fungi along the gradient of ecosystem development.
Identifying the drivers of microbial community composition in soil is challenging given the multitude of processes and the shifts in environmental conditions that occur at different scales in space and time (Fierer and Jackson, 2006). A robust approach to disentangle these drivers relies on the use of environmental chronosequences—which assumes a space for time replacement—and allows for large (or even multiple) time-scale analyses of favourable and constrained factors that influence community dynamics (Walker et al., 2010; Walker and Wardle, 2014; Dini-Andreote et al., 2015). The use of chronosequences intrinsically invokes the occurrence of ecological succession. Particularly for microbial ecologists, this requires a proper temporal calibration, that is, the establishment of a temporal framework that, for the modus operandi in microbial ecology, can vary from days to tens of years (Zhang et al., 2011; Voříšková et al., 2014). This is needed to effectively unravel how different mechanisms interact over time and at different scales, modulating the speed and outcome of ecological successions (Fierer et al., 2010; Walker and Wardle, 2014; Dini-Andreote et al., 2014, 2015).
The salt marsh at the island of Schiermonnikoog (The Netherlands) makes part of the few remaining pristine ecosystems in Western Europe (Bakker et al., 1997). This ecosystem has been formed through sand accumulation and progressive sedimentation of silt and clay particles, which resulted from cyclic tidal inundation. Together this cause a natural increase in the base of elevation of the marsh. Over time, this natural sedimentation causes the island to progressively extend eastwards (Olff et al., 1997; Van Wijnen et al., 1997). As a result, a natural chronosequence of ∼8 km length has developed from west to east, representing over a century of terrestrial ecosystem succession (Supplementary Figure S1; Olff et al., 1997). This environmental gradient has served as a natural laboratory for plant and animal ecology studies over the past 40 years (Bakker, 2014). The focus of previous works has been to understand the trophic and nontrophic community interactions (see, for example, Schrama et al., 2013) and the multilevel food-web dynamics (Schrama et al., 2012). Moreover, there is evidence for the contention that this chronosequence evolves by a gradual decoupling from the marine environment. In particular, early-soil sites are ‘allochthonous', being mostly fuelled by external input of nutrients and organisms (that is, the so-called ‘brown' food web). As succession proceeds, these influences are progressively curbed, and belowground nutrient dynamics is increasingly governed by the soil microbiota (that is, the so-called ‘green' food web) (Schrama et al., 2012).
Here, we make use of the Schiermonnikoog salt marsh chronosequence to investigate the patterns of fungal community succession. The major aim of the study was to assess how shifting environmental conditions drive fungal community assembly during soil colonization and further influence the spatiotemporal dynamics of these communities along a marine-to-terrestrial transition. In order to study the fungal community turnover in the system, we investigated the patterns of β-diversity in a multiscalar approach, that is, by examining the community temporal turnover within the successional stages (local; bimonthly scale) and across the stages over succession (regional; over 105 years). The objectives of this study were therefore: (1) to determine the patterns of fungal community size, composition and diversity along the salt marsh successional gradient; (2) to infer the key fungal ecophysiologies and adaptations along the chronosequence; (3) to infer from the fungal β-diversities within ‘local' and across ‘regional' scales the degree of multiscalar fungal community dynamics over time; and (4) to unravel how different abiotic factors influence the observed patterns of fungal β-diversity at both scales. We expected fungal community size and diversity patterns to follow the shifts in environmental productivity, that is, to increase over time and potentially stabilize at late successional stages. Fungal β-diversity at a local scale was expected to be driven mostly by shifts in edaphic properties and intrinsic seasonal dynamics within the sampling period (for example, plant phenology), thus reflecting differential community dynamics within the stages across succession. Finally, the regional patterns of fungal β-diversity were expected to closely mirror those observed for bacteria (Dini-Andreote et al., 2014), where community turnover is higher in early-successional soils and progressively decreases towards the late-successional stages. Taken together, this study enhances our understanding of the distribution and dynamism of soil fungal communities in the marine-to-terrestrial soil chronosequence, with special emphasis on the temporal patterns in fungal community succession and their potential role in ecosystem C dynamics.
Materials and methods
Study area and sample collection
Soil samples (n=60) were collected in May, July, September and November 2012 in a chronological transect located in the island of Schiermonnikoog N53o 30'E6o 10', The Netherlands. The study site presents a well-documented salt marsh chronosequence that has been calibrated using topographic maps, aerial photographs and monitoring permanent plots at different successional stages for over 20 years (Olff et al., 1997; Schrama et al., 2012). For this study, five successional stages were identified and estimated as 0, 5, 35, 65 and 105 years of succession (hereafter referred as ‘Stage' followed by the corresponding year) in May 2012 (Supplementary Figure S1). Triplicate plots (5 × 5 m2) were established at each successional stage (separated 25 m from each other) at the same base elevation (that is, position at the initial elevation gradient on the bare sand flats with a base elevation of 1.16 m±2.2 cm (mean±s.e.) above Dutch ordinance level). Importantly, differences in the base elevation reflect differences in inundation regimes, and therefore have strong influences on the dynamics and the fate of succession (Olff et al., 1997). In each plot, soil samples were collected by randomly taking 20 soil cores (5 cm diameter, 10 cm depth) using a sterile sampling device. Collected soil cores were pooled per plot in a sterile plastic bag, sealed and transported to the laboratory (<24 h). Soil cores were sieved (4 mm mesh size) to remove plant debris and to increase sample homogenization. Sieving and sample processing were conducted under sterile conditions to avoid contamination by airborne fungi. Homogenized soil samples per plot (n=60) were stored at −20 oC for total DNA extraction and at 4 oC for physicochemical analysis. In this study, no spatial variations within plots were considered. Variations within successional stages were addressed through triplicated plots (spatial) and across sampling times (temporal).
Soil DNA extraction and quantification
Total soil DNA was extracted from 0.5 g of initial material using the MoBio PowerSoil DNA isolation kit (MoBio Laboratories, Carlsbad, CA, USA). The manufacturer's protocol was slightly modified by the addition of glass beads (diameter 0.1 mm; 0.25 g) to the soil slurries followed by three cycles of bead beating (mini-bead beater, BioSpec Products, Bartlesville, OK, USA) for 60 s. Extracted DNA samples were quantified using the Quant-iT PicoGreen dsDNA assay kit (Invitrogen, Carlsbad, CA, USA) on a TECAN infinite M200 Pro (Maennedorf, Switzerland) plate reader reading at 485 nm excitation and 530 nm emission.
Quantitative PCR of the ITS region of the fungal rRNA gene
Quantitative PCR was used to calculate the number of fungal internal transcribed spacer (ITS) copies per g of dry weight soil in each sample, providing a proxy for fungal community size. Caution is warranted in terms of directly relating ITS copy numbers to fungal abundances or assuming that the obtained values are absolute quantifications of community size, as the number of ITS copies varies across different fungal lineages (Bellemain et al., 2010; Longo et al., 2013). Reactions were performed in 25 μl containing 12.5 μl Power SYBR Green PCR Master Mix (Applied Biosystems, Frankfurt, Germany), 1.25 μl of 20 mg ml−1 bovine serum albumin (Roche Diagnostics GmbH, Mannheim, Germany), 1.25 μl of each 10 μM primer (ITS1f 5′-TCCGTAGGTGAACCTGCGG-3′ and 5.8S 5′-CGCTGCGTTCTTCATCG-3′) (Gardes and Bruns, 1993; Vilgalys and Hester, 1990 respectively) and 10 ng of the sample DNA. Quantifications were carried out twice (technical replicates) for each soil sample and obtained values were averaged. The thermal cycling protocol was 95 oC for 10 min, 40 cycles of 95 oC for 1 min, 53 oC for 30 s and 72 oC for 1 min on the ABI Prism 7300 Cycler (Applied Biosystems). A standard curve was generated over six orders of magnitude from 102 to 107 copies of template per essay (R2=0.97) using a plasmid containing a cloned ITS region from Rhizoctonia solani that amplifies 318 bp. The quantitative PCR efficiency (E) was calculated according to the equation E=[10(−1/slope)−1]. The specificity of the amplification products was confirmed by melting curve analysis. Possible inhibitory effects of co-extracted humic compounds were checked by spiking samples with a range of known concentrations of the plasmid. No apparent inhibition was observed. Data are shown as log copy numbers per g of dry weight soil.
ITS amplification and Illumina MiSeq high-throughput sequencing
Amplification of the ITS1 region of the rRNA gene was performed twice per sample in 25 μl PCR reactions containing 10 μl of PRIME HotMasterMix (5 PRIME, Gaithersburg, MD, USA), 0.5 μl of each 10 μM primer and 10 ng of the sample DNA. The thermal cycling protocol was 94 oC for 3 min, 35 cycles of 94 oC for 45 s, 50 oC for 1 min, 72 oC for 1.5 min, followed by a 10-min final extension at 72 oC. We used the primer set ITS1f (Gardes and Bruns, 1993) and ITS2 (White et al., 1990), slightly modified for sequencing using the Illumina MiSeq platform (for a detailed description see Smith and Peay, 2014). PCR products were individually checked using gel electrophoresis and cleaned using the Agencourt Ampure XP kit (Beckman Coulter, Brea, CA, USA). Cleaned PCR products were pooled at equimolar concentrations (including replicated PCR products per sample) and sequenced in a paired-end (2 × 250 bp) Illumina MiSeq run at the Argonne National Laboratory in the Next Generation Sequencing Core (NGS).
DNA sequence analysis
The raw sequence data were demultiplexed using the QIIME toolkit (Caporaso et al., 2010). As the highly variable ITS amplicon length precludes paired-end assembly, only one of the reads (R1) was used for the analysis. Initial read quality filtering and dereplication were carried out using USEARCH v7 (Edgar, 2013) (USEARCH parameter: maxEE: remove reads with expected number of base call errors exceeding 0.5). We further used ITSx 1.0.11 (Bengtsson-Palme et al., 2013) to remove the flanking 5.8S and 18S rRNA gene sequences for optimal resolution of ITS1 clustering and removal of compromised and nonfungal target sequences. The extracted sequences were trimmed to equal lengths and further binned into operational taxonomic units (OTUs) at 97% identity using UPARSE (Edgar, 2013). Chimeric sequences were identified and removed using uchime_ref on USEARCH v7 (Edgar, 2013) against the UNITE UCHIME reference data set (version no. 7, release date 2015-03-11) (Abarenkov et al., 2010). Further analyses were carried out using QIIME (Caporaso et al., 2010). Taxonomy was assigned to each representative sequence using BLAST searches against the UNITE taxonomy string of the best matching UNITE sequence—UNITE ITS-reference database (version no. 7, release date 2015-03-02) (Abarenkov et al., 2010). For all OTU-based analyses, the original OTU table was filtered to contain only sequences taxonomically affiliated to fungi and further rarefied to a depth of 5500 sequences per sample (the fewest in a single sample) to minimize any effects of sampling effort on the analyses. The detailed ITS pipeline is available on http://brmicrobiome.org (Pylro et al., 2014). Sequences were deposited in the Qiita database (publicly available through study ID 10311) (qiita.ucsd.edu), and in the European Bioinformatics Institute (EBI) database (accession number ERP013288). To assign putative ecological functions to the fungal OTUs, we used more restrictive taxonomic affiliations. For that, each OTU was assigned to its closest genus available in UNITE using the SEED pipeline (Větrovský and Baldrian, 2013) and allocated into fungal ecophysiological categories (that is, saprotroph, plant pathogen, animal parasite, AM, ectomychorriza (EC)) based on published literature. The definition of the categories was the same as in Tedersoo et al. (2014). Fungal OTUs that were not assigned to any genus with known ecophysiology and those that were assigned to a genus with unclear ecophysiology were grouped into the ‘unassigned' category (Figure 3a).
It is worth mentioning that, although the ITS region has been broadly accepted as a universal barcode marker for fungi (Schoch et al., 2011), the selection of primer sets targeting either ITS1 or ITS2 goes with differential amplification biases against specific fungal lineages, in particular against those of Glomeromycota (Větrovský et al., 2015) and Chytridiomycota (Lindahl et al., 2013). In particular, within the phylum Glomeromycota there is the effect of homoplasy (that is, correspondence between parts arising from evolutionary convergence), where the same individual can carry divergent copies of ITS sequences. Moreover, the precision of the taxonomic assignment is subjected to the quality of data available in the database. This is also dependent on the length of the amplified fragment and on the OTU clustering method. Lastly, the current taxonomic resolution in the available databases is, to a large extent, biased due to the higher culturing effort devoted to strains involved in pathogenic interactions and/or industrial applications and less to marine fungal lineages. Collectively, these are key methodological constrains that must be acknowledged and further considered in ecological data interpretation. For a detailed user guide to fungal community assessment using high-throughput sequencing analysis, see Lindahl et al. (2013).
Statistical analysis
Estimates of α-diversity were based on an evenly rarefied OTU abundance matrix and included observed richness Sobs, evenness Evar (equitability; 1-complete equitability) and estimated diversity H' via the Shannon index. Sampling effort was estimated using Good's coverage. Statistical differences of measured α-diversity metrics across samples were determined using a nonparametric t-test with 999 Monte Carlo permutations and Bonferroni's correction.
Fungal β-diversity (that is, difference in community structure between samples) was measured using Bray–Curtis similarities calculated based on rarefied and square root transformed OTU abundances. All measured soil physicochemical properties were checked for normality using the Shapiro–Wilk test and further log(x+1) transformed—with the exception of pH—to improve normality and homoscedasticity for multivariate statistical analysis. The correlation strength and significance between fungal community structure and soil physicochemical properties were determined using RELATE (a nonparametric Mantel type test) run with 999 permutations. We further used a nonparametric multivariate regression between normalized soil physicochemical properties and the OTU-based Bray–Curtis similarity matrix implemented as distance-based linear modelling (DistLM) (McArdle and Anderson, 2001) and run with 999 permutations. Models were built using a step-wise selection procedure and the adjusted R2 selection criterion. Importantly, because of the high collinearity of soil physicochemical properties in our system, marginal DistLM was first used to determine which individual properties accounted for the greatest proportion of fungal community structure. We further used BEST to select for the best combination of less collinear factors—previously selected by marginal DistLM—to be incorporated into the models using the ‘forward' procedure within DistLM. The selected soil properties were subsequently used to build a constrained ordination plot using the best-fitted model in a distance-based redundancy analysis (Legendre and Anderson, 1999). These analyses, including the unconstrained principal coordinate analysis, were conducted using PRIMER6 and PERMANOVA+ (PrimerE Ltd, Ivybridge, UK).
Temporal turnover in fungal β-diversity within each stage of succession was determined via permutational multivariate analysis of variance (PerMANOVA; Anderson, 2001) using PRIMER6 and PERMANOVA+. PerMANOVAs were performed using sampling month as the main factor, allowing for full permutation of the raw data with Monte Carlo tests accounting for type III error, where the fixed effects sum to zero with 999 permutations. Changes in β-diversity within sites across different sampling time (using May as the baseline) were calculated as the difference, in percentage, between the average monthly Bray–Curtis similarities.
Results
Physicochemical properties of soils along the salt marsh chronosequence
The soil physicochemical properties varied significantly across the successional gradient (Supplementary Table S1). Briefly, the two early-successional stages (stages 0 and 5) revealed higher sand content than the later stages (92.3±0.5% and 92.1±0.1) and above-neutral pH values (averages of 8.7 and 8.3). The chronosequence evolves through progressive and intermittent sedimentations that increase the accretion of silt and clay particles over time (ranging from initially 2.7% silt and 5.0% clay at stage 0 up to 49.0% and 37.7%, respectively, at stage 105). The early-successional stages were sparsely vegetated soils with relatively low nutrient contents (soil organic matter (SOM), N-NH4+, N-NO3− and S-SO42–) and salinity levels. With succession, the soil physical structure, the dominant vegetation and other soil properties showed progressive shifts. Values of pH decreased to 7.4±0.1 at late stages (stages 65 and 105), whereas nutrient content and salinity increased, owing to an accretion and accumulation effect (Dini-Andreote et al., 2014, 2015). For detailed description of the metadata, see Supplementary Table S1.
Fungal ITS copy abundance and α-diversity along the soil chronosequence
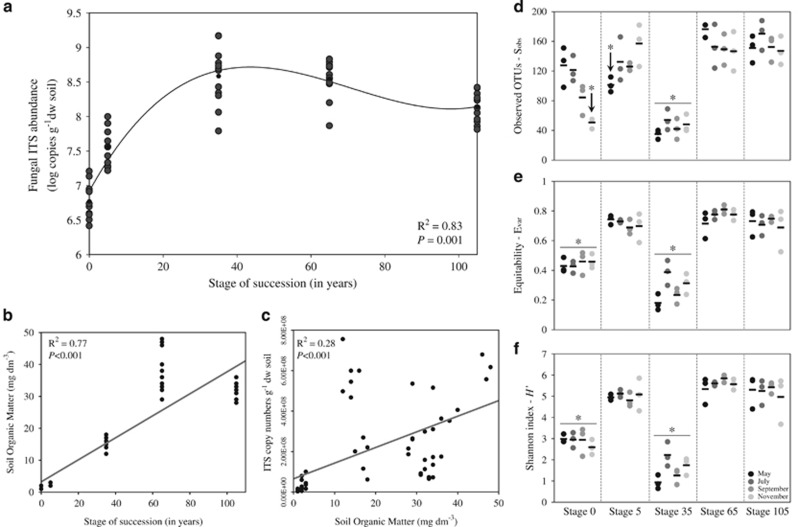
Quantifications of fungal ITS copy numbers were performed in duplicate with E=99.47% and R2=0.97. Within each stage, small but significant temporal variations in fungal ITS abundances were observed (Supplementary Figure S2). Along the successional gradient, a ∼10-fold increase occurred from stage 0 to 5 (average log values of 6.77 to 7.56; P<0.001), followed by an additional 10-fold increase between stages 5 and 35 (average log values of 7.56 to 8.58; P<0.001), remaining statistically stable thereafter. Cubic regression was used to model the fungal ITS abundance increments along the succession (ANOVA P=0.001, R2=0.83; Figure 1a). A strong and significant increase in the SOM values was found along the successional gradient (ANOVA P=0.001, R2=0.77; Figure 1b). Moreover, linear regression showed a significant (ANOVA P=0.001) effect of SOM as a modulator of soil fungal ITS abundance in the chronosequence (R2=0.28; Figure 1c).
Figure 1.
Fungal ITS copy abundance and α-diversity in the salt marsh chronosequence. (a) Cubic regression of fungal ITS abundances along the chronosequence—abundances are indicated by log copy numbers of the ITS region of the fungal rRNA gene per g of dry weight soil, using quantitative PCR. (b, c) Linear regressions displaying the increments in SOM along the salt marsh chronosequence (b), and the relationship between SOM and fungal ITS abundance (c). Statistics are provided on the panels. (d–f) Distributions of α-diversity measurements according to sampling time across successional stages: (d) observed OTUs (Sobs), (e) equitability—(Evar) and (f) Shannon index (H'). Comparisons of α-diversity across stages were carried out by nonparametric t-tests with 999 Monte Carlo permutations (P<0.001).
Estimates of fungal α-diversity were based on evenly rarefied OTU matrices (5500 sequences per sample). Sequence clustering yielded a total of 917 OTUs (116±49 per sample), representing a (Good's) coverage of 100% for all samples. The number of observed OTUs (Sobs) showed the highest within-stage dispersion values at the early-successional sites (stages 0 and 5). In fact, significant differences (P<0.001) were observed within stage across the sampling time (as indicated by black arrows; Figure 1d). Equitability (Evar) and Shannon diversity (H') indices did not differ within stages across sampling time. However, across stages, both Evar and H' showed lowered values at stages 0 and 35 (P<0.001), being statistically similar across the other successional sites (stages 5, 65 and 105) (Figures 1e and f). Notably, at the intermediate site (stage 35), we observed that one single OTU (taxonomically affiliated to the fungus Metarhizium anisopliae) accounted for 68.8% (averaged value) of the sequences, explaining the lowered α-diversity found at this stage.
Fungal community compositions and predicted ecophysiologies along the soil chronosequence
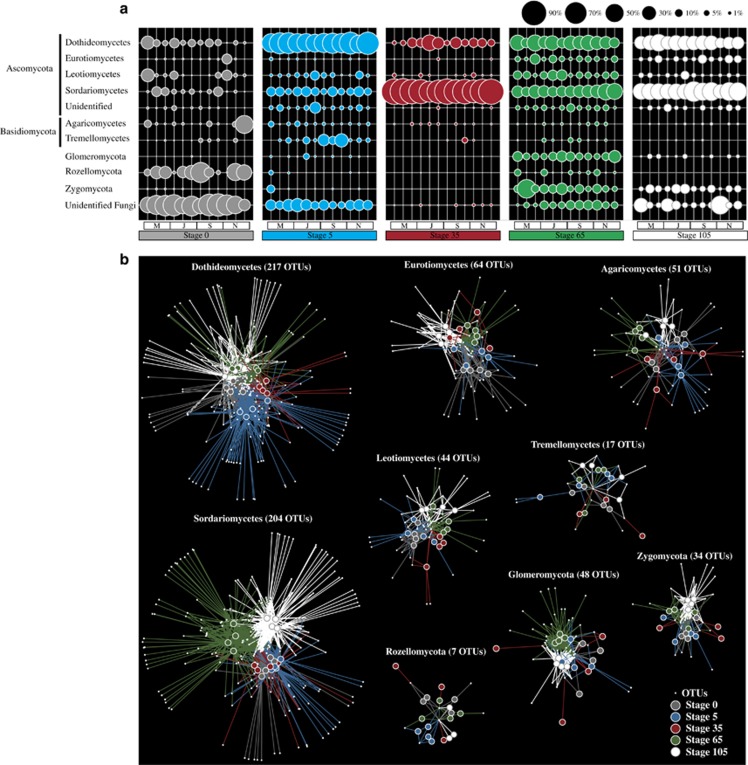
Fungal ITS sequences (917 OTUs) were distributed over 16 classes within 6 phyla. Across all successional stages, these belonged mainly to Dikarya (66.8% Ascomycota, with Basidiomycota accounting for only 4.3%). Dikarya were present in low abundance at stage 0, where sequences were mainly affiliated to Rozellomycota (20.7%). However, a large proportion of the sequences in this stage (45 OTUs, 49.5%) could not be affiliated to any fungal phylum (that is, the sequences were not affiliated to any close references in the database that are potentially related to basal fungal lineages, as previously reported (Baldrian et al., 2012)). At stage 5, the sequences were mostly affiliated with members of the Dothideomycetes (141 OTUs, 55.5%) and Sordariomycetes (81 OTUs, 9.7%), apart from other low-abundance groups—Tremellomycetes (6.4%), Leotiomycetes (3.5%), Agaricomycetes (1.8%), and Glomeromycota, Eurotiomycetes, Rozellomycota and Zygomycota (all <1%). Unexpectedly, stage 35 was mostly dominated (even across sampling time) by sequences that were closely affiliated to M. anisopliae (average value of 68.8%), apart from Dothideomycetes (65 OTUs, 12.2%), of which 42.5% of sequences within this group affiliated to Preussia polymorpha. At stage 65, the fungal community was composed of Dothideomycetes (24.9%), Sordariomycetes (22.1%), Zygomycota (12.4%), Glomeromycota (10.1%), Leotiomycetes (7.6%), Eurotiomycetes (5.1%) and Rozellomycota (2.2%), with Agaricomycetes and Tremellomycetes being <1%. Finally, stage 105 resembled stage 65, with small differences in OTU composition and in the relative abundances of specific taxonomic groups (Figures 2a and b). Here, the classes Sordariomycetes and Dothideomycetes were most abundant (39.9% and 26.7%, respectively), whereas low numbers of Glomeromycota (1.5%) were found. Figures 2a and b show the detailed data. Notably, the Chytridiomycota accounted for only a minor fraction of the fungal communities across all sites (<1% average values of 0.62%, not shown). Moreover, all sequences affiliated with the phylum Glomeromycota—a well-characterized phylum encompassing AM fungi—belonged to the class Glomeromycetes.
Figure 2.
Composition of soil fungal communities in the salt marsh chronosequence. (a) Relative abundances of fungal taxa at the phylum level, except for the phyla Ascomycota and Basidiomycota shown at the class level. Only taxonomic groups that sum >1% across all samples are shown. (b) Networks built per fungal taxonomic group showing shared OTUs among different stages of succession. The edge-weighted spring-embedded algorithm pulled together systems with similar structures and OTUs with similar associations.
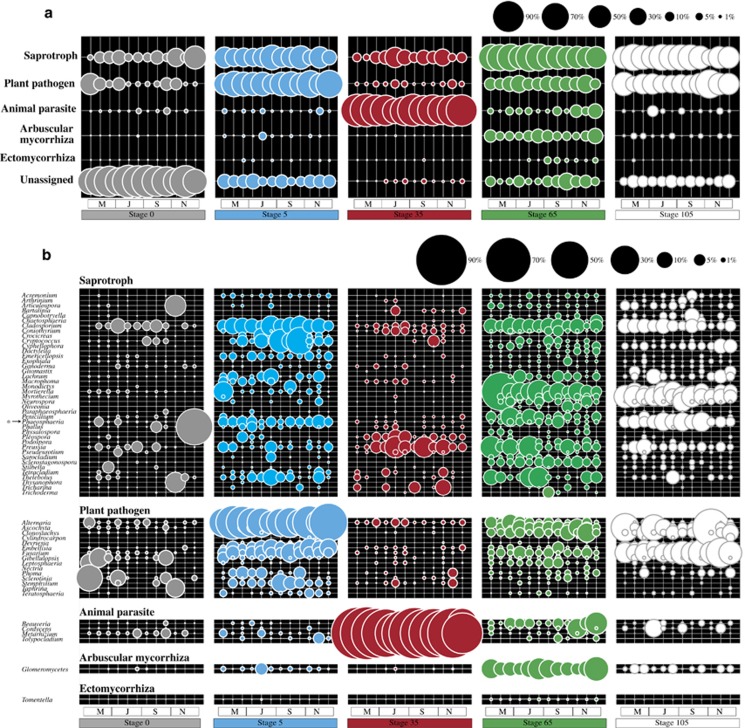
The assignment of sequences to fungal genera allowed an analysis of trends in the abundance of predicted fungal ecophysiologies along succession. First, stages 65 and 105, constituting well-vegetated sites with high amounts of SOM (average 35±5 g dm–3), revealed the highest proportions of fungi with predicted saprotrophic lifestyles (54±12% and 48%±11, respectively). This stood in sharp contrast to the early- and intermediate-successional stages, where such proportions were lower (stage 0: 14±12% stage 5: 34±10% and stage 35: 15±9%). The composition of the predicted saprotrophic fungi also shifted across the successional stages. These were mostly represented, at the early-successional stages, by genera such as Cladosporium, Cryptococcus and Phaeosphaeria; at intermediate stage by Preussia; and at the late stages by Mortierella, Myrothecium, Cladosporium, Preussia and Phaeosphaeria. Concerning the predicted plant-pathogenic fungi, the genera Alternaria, Fusarium, Ascochyta and Emellisia were the most abundant ones found across all successional stages. Concerning the predicted animal parasites or pathogens, the genus Metarhizium dominated. It peaked at stage 35, but was also present, at a low abundance (<2%), across other successional stages. Regarding mycorrhizal fungi, EC taxa accounted for only a small fraction of the identified fungi (<1%) across all stages. AM (in this case, all affiliated to Glomeromycetes) peaked at stage 65 (10±5%) and became lower in abundance at stage 105 (1.5±1%) (see Figures 3a and b for details).
Figure 3.
Distribution of predicted fungal ecophysiologies in soil samples along the salt marsh chronosequence. (a) Relative abundance distributions. Fungal OTUs that were not assigned to any genus with known ecophysiology or to a genus with unclear ecophysiology were grouped into the ‘unassigned' category (see Materials and methods for details). (b) Taxonomic composition of each ecophysiological category at the genus level (except for Glomeromycetes–unclassified genera). Only genera >1% in at least one sample are shown. OTUs that were not assigned to any ecophysiological category were omitted. The displayed genera and ecophysiological assignments represented different coverage values (%) of the total fungal communities in each successional stage, as follows: stage 0: 23±14, stage 5: 85±4, stage 35: 98±1, stage 65: 86±8 and stage 105: 87±4 (averaged mean values per stage±s.d.). For principal coordinate analysis (PCO) and additional boxplots of ecophysiological categories across soil successional stages, see Supplementary Figure S3. *Specific members within this genus have also been reported as plant pathogens (see Tedersoo et al., 2014 for details).
Differences in fungal community turnover within (local scale) and across (regional scale) successional stages and relation to environmental properties
Analyses of fungal community turnover at the local scale (that is, analysing within-stage β-diversity over time) revealed relatively high shifts in the communities at the early-successional stages (stage 0: Pseudo-F=2.038, P=0.001; stage 5: Pseudo-F=2.405, P=0.001). In contrast, the intermediate- (stage 35: Pseudo-F=1.600, P=0.101) and late-successional stages (stage 65: Pseudo-F=1.119, P=0.234; stage 105: Pseudo-F=1.035, P=0.416) did not vary significantly over time (Supplementary Table S2). The short-scale temporal community turnover at the early-successional stages did not significantly correlate (P>0.05) to any shifts in the measured soil physicochemical properties.
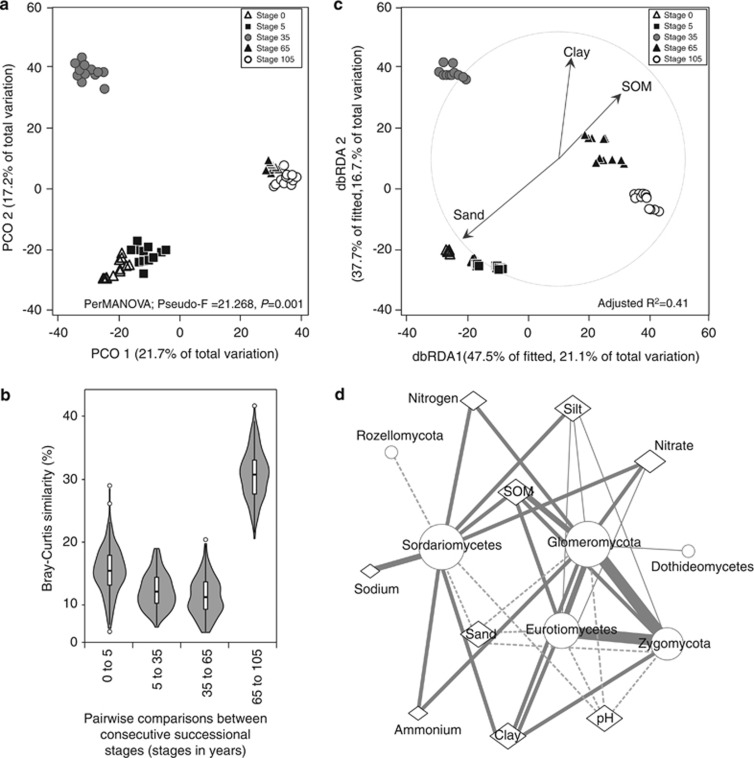
Analyses of fungal community turnover at the regional scale (that is, considering the between-stage β-diversity) revealed significant differences in the fungal communities along the successional gradient (Pseudo-F=21.268, P=0.001; Figures 4a and b; Supplementary Table S2). Overall, the community structure was significantly correlated with the soil physicochemical properties (RELATE ρ=0.578, P<0.001). Analyses of individual significant properties (P<0.001) by marginal DistLM resulted in the selection of sand (Pseudo-F=14.8, proportion=0.20), clay (Pseudo-F=13.5, proportion=0.19) and SOM (Pseudo-F=13.7, proportion=0.19) as the predictor variables that best modelled the regional β-diversity pattern. Collectively, these factors correlated significantly with the fungal community structures (BEST ρ=0.663, P<0.001). These properties were selected based on their high explanatory proportions in marginal DistLM analyses. Hence, other colinear properties that explained smaller proportions—despite being significant (P<0.001)—were not considered in the model (see Supplementary Table S3 for additional models including an increasing number of predictor variables). The best DistLM model is shown by distance-based redundancy analysis, explaining 37.8% of the total variation, with an adjusted R2 of 0.41 (Figure 4c).
Figure 4.
Patterns and drivers of fungal communities across successional stages in the salt marsh chronosequence. (a) Principal coordinate analysis (PCO) ordination based on Bray–Curtis similarities calculated on rarefied OTU abundances. Variance explained by each PCO axis is given in parentheses. (b) Distance-based redundancy analysis (dbRDA) plot illustrating the ‘best' fitted DistLM model (adjusted R2=0.414) containing forward selected predictor variables (P<0.001) for β-diversity. Axis legends include % of variation explained by the fitted model and % of total variation explained by the axis. (c) Violin plot displaying pairwise fungal community similarity (Bray–Curtis) analysed between consecutive successional stages. (d) Associations between soil physicochemical properties (diamonds) and fungal taxonomic groups (circles) visualized as a network. Thickness of lines represents stronger positive (solid line) or negative (dashed line) correlations. Only Spearman's rho >0.5 and statistically significant (P<0.01) are shown. Node sizes are proportional to the number of correlations. For a complete list of correlational values see Supplementary Tables S4 and S5.
The magnitude of effects of the individual soil physicochemical properties on fungal taxon distributions and the relationships within different taxonomic groups were further tested by Spearman correlations. These analyses revealed the classes Agaricomycetes, Tremellomycetes and Rozellomycota to be weakly (ρ<0.5) but significantly (P<0.01) correlated with the soil properties that prevailed at the early-successional stages (that is, sandy and nutrient-poor soils under high pH and daily seawater influences). On the other hand, Eurotiomycetes, Sordariomycetes, Glomeromycota and Zygomycota were correlated mostly with soil properties that prevailed at the late-successional stages (P<0.01), with a few strong correlations between each other (for instance, Glomeromycota–Zygomycota ρ=0.753, P<0.01; Zygomycota–Eurotiomycetes ρ=0.784, P<0.01; Figure 4d). For a full list of pairwise correlational values see Supplementary Tables S4 and S5.
Discussion
This study provides the first high-resolution assessment of fungal communities in salt marsh soils by elucidating the differential patterns in community composition and assessing the shifts in the fungal α- and β-diversities in an evolving ecosystem. We addressed the relatedness between the observed temporal changes and the shifting environmental conditions, aiming to understand how fungal communities establish in the chronosequence and may affect local ecological processes such as C transformations. As salt marshes are important C sinks, understanding the distribution and drivers of fungal communities in these threatened areas are of critical importance (Deegan et al., 2012; Kirwan and Mudd, 2012). Thus, the results of this study may directly affect ecological models that aim to predict ecosystem responses to climate change and direct anthropogenic perturbations (see Bardgett et al., 2008; Chapin et al., 2009).
Fungal communities along the marine-to-terrestrial transition
In contrast to other studies in salt marshes that have focussed on more mature soil sites (see, for example, Buchan et al., 2002; Torzilli et al., 2006), we aimed to understand the dynamic changes of fungal communities across a consistently changing landscape. Together, the study sites represent a natural marine-to-terrestrial transition gradient, which allow for the investigation of fungal community succession and adaptation. We first quantified a proxy for fungal community size and found that fungal ITS abundances increased 10- to 100-fold from the early- to intermediate/late-successional stages (Figure 1a). This progressive increase was partially explained by the correlation to the gradual increments in SOM (Figures 1b and c), possibly indicating that after a certain level of SOM, influences on fungal communities may level off. The patterns of fungal ITS copy number increments along the chronosequence appeared to be similar to primary succession observed in coal mine deposits where—in this case—fungal biomass also increased with successional stage but peaked at an intermediate stage and decreased later (Baldrian et al., 2008). However, one important caveat of our approach relates to the fact that, as communities significantly changed in composition across successional stages (Figure 4a) and the number of ITS copies varies across different fungal taxa (Bellemain et al., 2010; Longo et al., 2013), our measurements of community size (that is, ITS copy numbers) demand a careful interpretation. As such, these data must be considered as indicative of potential shifts in fungal community sizes along succession rather than as absolute values. Contrary to initial expectations, and despite the 10- to 100-fold reduced fungal ITS abundances, the early-successional sites (with the exception of one sampling time at stage 0) exhibited species richnesses comparable to those of more mature soils (Figures 1d–f). Despite the unexpected high richnesses, a large proportion of OTUs from stage 0 matched as-yet-unclassified taxa, constraining any inference about their ecophysiology (Figures 2a and 3a). Of those taxonomically assigned, most belonged to the Rozellomycota, a basal clade within the Fungi (James et al., 2006a, 2006b) that contains uniflagellated and unwalled zoospores (Corsaro et al., 2014). Thus, ready motility by swimming might give an eco-evolutionary asset to the fungi dwelling at these intermittently flooded sites. As such, it is tempting to speculate that the majority of the novel unidentified sequences belong to fungal lineages that are potentially affiliated to an early radiation of the Dikaria clade, close to the chytrid branches (Richards et al., 2012) and/or to yeast-like basal fungal lineages. These arguments collectively corroborate previous studies that have suggested that coastal environments host diverse as-yet-unclassified fungal lineages that may be typical for the (ancient) transition from marine to terrestrial systems (Le Calvez et al., 2009; Mohamed and Martiny, 2011).
As succession proceeds to the more mature terrestrial stages, the fungal communities apparently evolved towards a dominance of filamentous and, to a larger extent, saprotrophic fungi, as their mode of exploration for nutrients is fine tuned to the utilization of complex substrates such as the organic matter derived from plant biomass (Richards et al., 2012; Treseder and Lennon, 2015). As primary agents of plant litter decomposition, saprotrophic fungi produce hyphal networks throughout the soil matrix that promote organic matter-derived nutrient distribution through dynamic channels (Crowther et al., 2012). Interestingly, we found the types of putative saprotrophic fungi to shift significantly along the chronosequence (Figure 3b). Such shifts seem to be determined by the sources of organic materials at each successional stage that are reflective of the local vegetation (Urbanová et al., 2015). Fungal lineages have the ability to directly or indirectly access these materials as nutrients, and this has likely led to the creation of a variable environmental filter along the gradient. Known as effective decomposers that boost C mineralization processes, saprotrophic fungi might also play a role in the deposition and accretion of recalcitrant C in these soils, an important factor accounting for long-term C storage (Treseder and Lennon 2015).
Fungi with a (beneficial) plant-associated lifestyle, that is, AM and EC, were found as only small fractions of the retrieved sequences, primarily at stage 65. This stage has previously been shown to have a peak in plant diversity (Schrama et al., 2012). The extent of mycorrhizal colonization of the plants occurring at the study sites was not explored. However, the dominant plant taxa at stages 5 and 105, Spergularia media and Elytrigia atherica, are nonmycorrhizal, similar to Juncus gerardii. In contrast, several of the dominant plant species of stages 35 and 65 (Limonium vulgare, Aster tripolium, Festuca rubra and Artemisia maritima) are known to form AM symbioses with Glomeromycota (Supplementary Table S1; Wilde et al., 2009; Druva-Lusite and Ievinsh, 2010). The highest prevalence of the AM fungi at the stage 65 may therefore be related to the highest diversity of AM-forming dominants, or it may indicate a phosphorous deficiency that promotes AM formation.
In contrast to other studies (see, for example, Blaalid et al., 2012; Brown and Jumpponen, 2014), we did not observe a clear directional shift in the fungal communities across successional stages. This was mostly because of the extreme abundance of the OTU affiliated with M. anisopliae at stage 35, which may indicate a high abundance of either the hyphal or spore forms. M. anisopliae is a well-known entomopathogenic fungus (ecologically assigned as ‘animal parasite'), but a saprotrophic and/or plant-associated lifestyle is also possible (Meyling and Eilenberg, 2007). We advocate further studies to properly confirm the identity (for example, by isolation and culture) and ecological function of this organism, as well as the factors mediating its high abundance at the intermediate soil stage. Clearly, the dominance of this OTU significantly changed patterns of community richness and diversity at this stage (Figures 1d–f), which potentially exerted an effect on the ecological succession trajectory.
Understanding fungal community turnover at local and regional scales is critical to explain ecological succession
Microbial ecologists have only recently started to investigate the patterns of microbial community dynamics in natural systems (see, for example, Nemergut et al., 2007; Blaalid et al., 2012; Brown and Jumpponen, 2014; Dini-Andreote et al., 2014). As changes in microbial community composition are often associated with variations in the functional capabilities of the communities (Fierer et al., 2007; Salles et al., 2009), unravelling the temporal turnover patterns is crucial for understanding ecosystem processes. Here, we first hypothesized that local scale β-diversity is mostly driven by (shifts in) edaphic properties and intrinsic seasonal dynamics. Indeed, at the early-successional sites, a great turnover in species composition was found that was not related to any shifts in soil properties across sampling time. Consistent with previous data (Dini-Andreote et al., 2014, 2015) for bacterial communities, the high fungal β-diversity may have been governed by stochastic processes. Supporting this assertion are the facts that (1) the early-successional sites are dynamic, and experience daily cycles of inundation that contribute to fluctuations in local environmental conditions (for example, salinity, oxygen content and temperature) (Dini-Andreote et al., 2014; Supplementary Figure S1c); and (2) there is random immigration and interim establishment of organisms at these ‘allochthonous' soil sites, as reflected in the differences in α-diversity (Sobs) across sampling time (Figure 1d). Moreover, the weak correlations found within fungal taxonomic groups and between fungal taxa with local soil properties also provide support for the potentially weak selection and interim establishment of organisms at these sites (Figure 4d, Supplementary Tables S4 and S5). As succession proceeds, the local β-diversity stabilizes, and does not differ across samplings within the intermediate- and late-successional stages. These findings were partially in contrast with our initial hypothesis, as intrinsic seasonal dynamics (in this case, plant phenology and seasonality) was expected to exert significant effects on the fungal community turnover, as verified in other ecosystems (see, for example, Zinger et al., 2009; Voříšková et al., 2014). An underlying cause of this finding could be that most of the degraded plant litter on the salt marsh comes from dead rather than fresh plant material (Torzilli et al., 2006). This could, in turn, limit the role of plant seasonality as a temporal driver of the fungal community turnover in salt marshes.
Regional-scale analyses of the fungal β-diversity revealed a clear segregation of the communities across the different successional stages (Supplementary Table S2), either in the community compositions, as indicated in the β-diversity analyses (Figure 4a), or in the succession rates, as shown by pairwise community similarity comparisons (Figure 4b). The latter closely approximated our expectation, as community dissimilarity decreased towards the late soil sites, as previously shown for bacteria (Dini-Andreote et al., 2014). Furthermore, a detailed meta-analysis revealed that not only the β-diversity patterns of fungal and bacterial communities resembled each other across scales, but also that the progressive increments in bacterial 16S rRNA and fungal ITS abundances occur at similar rates (R2=0.76, P<0.001; Supplementary Figure S4). This finding concurs with data from previous studies (Baldrian et al., 2008; Brown and Jumpponen, 2014) but contrasts with those in another one (Ohtonen et al., 1999), suggesting that a shift from bacterial- to fungal-dominated systems occurred concomitantly with vegetation establishment. In such conditions, fungi might start to outcompete bacteria, as they may more effectively utilize the carbon available from plants or, alternatively, they establish close (mycorrhizal) associations with plants. In our study, however, although we did not use a direct measurement of biomass (see Materials and methods for details), we did obtain evidence to suggest that bacterial and fungal communities were constrained in a similar manner following the environmental productivity.
Moreover, we effectively modelled the fungal β-diversity at the regional scale by integrating soil physical structure and SOM as predictive variables (Figure 4c). One intriguing interpretation is the contention that nutrient-poor and barely vegetated ‘nascent' soils are primarily under ‘allochthonous' control that are ‘erratic' and thus incites dynamic shifts in community composition. At the other end of the spectrum, the late stages of the soil chronosequence (mostly under ‘autochthonous' control) are less amenable to strong (temporal or even spatial) shifts in community composition. We suggest that, at intermediate and late soil sites, the local plant species have a major role in modulating the SOM, particularly by the progressive accretion of C-rich sources (that is, root exudation and litter deposition). Thus, the natural dynamics of plant colonization along the salt marsh successional gradient is likely to modulate the belowground SOM, which is directly intertwined with shifts in soil fungal communities. The influence of SOM on soil fungal communities has been previously reported (Hartmann et al., 2009; Millard and Singh, 2010); not only the quantity but also the quality status and turnover rate of SOM were found to influence specific fungal populations along an ecosystem landscape (Zinger et al., 2011). Yet, in our study, it appeared that, at a local scale, the magnitude of the shifts in SOM quantity and quality did not exceed the effects imposed by the dominant vegetation, resulting in a lowered fungal turnover within sites. On the other hand, the dynamic shifts in fungal communities at the regional scale revealed a progressive stabilization towards late stages, in which communities are driven towards equilibrium with the local environment. In such ‘mature soil' sites, a constant flux of carbonaceous compounds may be feeding a presumably strong network of interactive fungal types.
Conclusions and perspectives
There is a myriad of ways by which microbial metabolic activities can influence soil–atmosphere C exchanges (Bardgett et al., 2008). Importantly, a better understanding of the organisms as well as the biotic/abiotic mechanisms involved in C dynamics is needed to efficiently predict responses to environmental perturbations such as anthropogenic and climate change impacts. Following this line of reasoning, this study has two important implications. First, the investigation of an undisturbed salt marsh chronosequence provided a unique opportunity to unravel how fungal communities evolve in a marine-to-terrestrial ecosystem gradient. In particular, this highlights the interwovenness of the mechanisms that underlie fungal community assembly and the temporal dynamics therein. Second, as fungal ecophysiology is critical for ascertaining the processes mediating the (fungal-driven) C dynamics in soil (McGuire and Treseder, 2010), integrating ecophysiological inference into an ecological succession framework is important. It provides the basis for the development of theoretical models that link shifts in community structures to the roles of community members in processes like C dynamics. Future studies should cover a broader spectrum of salt marshes, looking for idiosyncrasies and commonalities, in an effort to create a holistic view of fungal roles on ecosystem succession and functioning. The progressive integration of information in conjunction with the increasing resolution of the methods for fungal taxonomic and ecophysiological annotation may ultimately allow us to accurately quantify and model the role of fungal community members in salt marsh C budgets.
Acknowledgments
We thank the ‘Nederlandse Vereniging voor Natuurmonumenten' for granting us access to the salt marsh. This research was supported by the Netherlands Organisation for Scientific Research (NWO). Petr Baldrian was supported by the Institutional Research Concept of the Institute of Microbiology of the CAS, Prague (RVO61388971). We thank Rogério B Lopes for insightful discussions and Angelique Gobet and Emil Ruff for kindly sharing an R script. We also thank Maria Julia L Brossi and Cyrus A Mallon for technical support and critical reading of the manuscript.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
Supplementary Material
References
- Abarenkov K, Henrik Nilsson R, Larsson KH, Alexander IJ, Eberhardt U, Erland S et al. (2010). The UNITE database for molecular identification of fungi – recent updates and future perspectives. New Phytol 186: 281–285. [DOI] [PubMed] [Google Scholar]
- Anderson MJ. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol 26: 32–46. [Google Scholar]
- Bakker JP. (2014) Ecology of Salt Marshes: 40 Years of Research in the Wadden Sea. Wadden Academy Ruiterskwartier: Leeuwarden, The Netherlands. [Google Scholar]
- Bakker JP, Esselink P, van der Wal R, Dijkema KS. (1997). Options for restoration and management of coastal salt marshes in Europe. In: Urbanska KM, Webb NR, Edwards PJ (eds), Restoration Ecology and Sustainable Development. Cambridge University Press: Cambridge, UK, pp 286–322. [Google Scholar]
- Baldrian P, Kolařík M, Štursová M, Kopecký J, Valášková V, Větrovský T et al. (2012). Active and total microbial communities in forest soil are largely different and highly stratified during decomposition. ISME J 6: 248–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldrian P, Trögl J, Frouz J, Šnajdr J, Valášková V, Merhautová V et al. (2008). Enzyme activities and microbial biomass in topsoil layer during spontaneous succession in spoil heaps after brown coal mining. Soil Biol Biochem 40: 2107–2115. [Google Scholar]
- Bardgett RD, Freeman C, Ostle NJ. (2008). Microbial contributions to climate change through carbon cycle feedbacks. ISME J 2: 805–814. [DOI] [PubMed] [Google Scholar]
- Bellemain E, Carlsen T, Brochmann C, Coissac E, Taberlet P, Kauserud H. (2010). ITS as an environmental DNA barcode for fungi: an in silico approach reveals potential PCR biases. BMC Microbiol 10: 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengtsson-Palme J, Ryberg M, Hartmann M, Branco S, Wang Z, Godhe A et al. (2013). Improved software detection and extraction of ITS1 and ITS2 from ribosomal ITS sequences of fungi and other eukaryotes for analysis of environmental sequencing data. Methods Ecol Evol 4: 914–919. [Google Scholar]
- Blaalid R, Carlsen T, Kumar S, Halvorsen R, Ugland KI, Fontana G et al. (2012). Changes in the root-associated fungal communities along a primary succession gradient analysed by 454 pyrosequencing. Mol Ecol 21: 1897–1908. [DOI] [PubMed] [Google Scholar]
- Brown SP, Jumpponen A. (2014). Contrasting primary successional trajectories of fungi and bacteria in retreating glacier soils. Mol Ecol 23: 481–497. [DOI] [PubMed] [Google Scholar]
- Buchan A, Newell SY, Moreta JI, Moran MA. (2002). Analysis of internal transcribed spacer (ITS) regions of rRNA genes in fungal communities in a southeastern U.S. salt marsh. Microb Ecol 43: 329–340. [DOI] [PubMed] [Google Scholar]
- Burke DJ, Hamerlynck EP, Hahn D. (2002). Interactions among plant species and microorganisms in salt marsh sediments. Appl Environ Microbiol 68: 1157–1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapin FS, McFarland J, McGuire AD, Euskirchen ES, Ruess RW, Kielland K. (2009). The changing global carbon cycle: linking plant-soil carbon dynamics to global consequences. J Ecol 97: 840–850. [Google Scholar]
- Chmura GL, Anisfeld SC, Cahoon DR, Lynch JC. (2003). Global carbon sequestration in tidal, saline wetland soils. Glob Biogeochem Cy 17: 1111–1133. [Google Scholar]
- Corsaro D, Walochnik J, Venditti D, Steinmann J, Müller KD, Michel R. (2014). Microsporidia-like parasites of amoebae belong to the early fungal lineage Rozellomycota. Parasitol Res 113: 1909–1918. [DOI] [PubMed] [Google Scholar]
- Crowther TW, Boddy L, Hefin Jones T. (2012). Functional and ecological consequences of saprotrophic fungus–grazer interactions. ISME J 6: 1992–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deegan LA, Johnson DS, Warren RS, Peterson BJ, Fleeger JW, Fagherazzi S et al. (2012). Coastal eutrophication as a driver of salt marsh loss. Nature 490: 388–392. [DOI] [PubMed] [Google Scholar]
- Dini-Andreote F, Pereira e Silva MC, Triadó-Margarit X, Casamayor EO, van Elsas JD, Salles JF. (2014). Dynamics of bacterial community succession in a salt marsh chronosequence: evidences for temporal niche partitioning. ISME J 8: 1989–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dini-Andreote F, Stegen JC, van Elsas JD, Salles JF. (2015). Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc Natl Acad Sci USA 112: E1326–E1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Druva-Lusite I, Ievinsh G. (2010). Diversity of arbuscular mycorrhizal symbiosis in plants from coastal habitats. Environ Exp Biol 8: 17–34. [Google Scholar]
- Edgar RC. (2013). UPARSE: Highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10: 996–998. [DOI] [PubMed] [Google Scholar]
- Fierer N, Bradford MA, Jackson RB. (2007). Toward an ecological classification of soil bacteria. Ecology 88: 1354–1364. [DOI] [PubMed] [Google Scholar]
- Fierer N, Jackson RB. (2006). The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA 103: 626–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Nemergut D, Knight R, Craine JM. (2010). Changes through time: integrating microorganisms into the study of succession. Res Microbiol 161: 635–642. [DOI] [PubMed] [Google Scholar]
- Gardes M, Bruns TD. (1993). ITS primers with enhanced specificity for basidiomycetes –application to the identification of mycorrhizae and rusts. Mol Ecol 2: 113–118. [DOI] [PubMed] [Google Scholar]
- Hartmann A, Schmid M, van Tuinen D, Berg G. (2009). Plant-driven selection of microbes. Plant Soil 321: 235–257. [Google Scholar]
- Heimann M, Reichstein M. (2008). Terrestrial ecosystem carbon dynamics and climate feedbacks. Nature 451: 289–292. [DOI] [PubMed] [Google Scholar]
- Heinemeyer A, Hartley IP, Evans SP, De la Fuente JAC, Ineson P. (2007). Forest soil CO2 flux: uncovering the contribution and environmental responses of ectomycorrhizas. Glob Change Biol 13: 1786–1797. [Google Scholar]
- Hildebrandt U, Janetta K, Ouziad F, Renne B, Nawrath K, Bothe H. (2001). Arbuscular mycorrhizal colonization of halophytes in Central European salt marshes. Mycorrhiza 10: 175–183. [Google Scholar]
- Högberg P, Nordgren A, Buchmann N, Taylor AF, Ekblad A, Högberg MN et al. (2001). Large-scale forest girdling shows that current photosynthesis drives soil respiration. Nature 411: 789–792. [DOI] [PubMed] [Google Scholar]
- James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ et al. (2006. a). Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443: 818–822. [DOI] [PubMed] [Google Scholar]
- James TY, Letcher PM, Longcore JE, Mozley-Standridge PD, Powell MJ, Griffith GW et al. (2006. b). A molecular phylogeny of the flagellated fungi (Chytridiomycota) and description of a new phylum (Blastocladiomycota). Mycologia 98: 860–871. [DOI] [PubMed] [Google Scholar]
- Kirwan ML, Mudd SM. (2012). Response of salt-marsh carbon accumulation to climate change. Nature 489: 550–553. [DOI] [PubMed] [Google Scholar]
- Le Calvez T, Burgaud G, Mahe S, Barbier G, Vandenkoornhuyse P. (2009). Fungal diversity in deep-sea hydrothermal ecosystems. Appl Environ Microbiol 75: 6415–6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre P, Anderson M. (1999). Distance-based redundancy analyses: testing multispecies responses in multifactorial ecological experiments. Ecol Monogr 69: 1–24. [Google Scholar]
- Lindahl BD, Nilsson RH, Tedersoo L, Abarenkov K, Carlsen T, Kjøller R et al. (2013). Fungal community analysis by high-throughput sequencing of amplified markers — a user's guide. New Phytol 199: 288–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo AV, Rodriguez D, da Silva Leite D, Toledo LF, Almeralla CM, Burrowes PA et al. (2013). ITS1 copy number varies among Batrachochytrium dendrobatidis strains: implications for qPCR estimates of infection intensity from field-collected amphibian skin swabs. PLoS One 8: e59499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArdle B, Anderson M. (2001). Fitting multivariate models to community data: a comment on distance-based redundancy analysis. Ecology 82: 290–297. [Google Scholar]
- McGuire KL, Treseder KK. (2010). Microbial communities and their relevance for ecosystem models: decomposition as a case study. Soil Biol Biochem 42: 529–535. [Google Scholar]
- Meyling NV, Eilenberg J. (2007). Ecology of the entomopathogenic fungi Beauveria bassiana and Metarhizium anisopliae in temperate agroecosystems: potential for conservation biological control. Biol Control 43: 145–155. [Google Scholar]
- Millard P, Singh B. (2010). Does grassland vegetation drive soil microbial diversity? Nutr Cycl Agroecosys 88: 147–158. [Google Scholar]
- Mohamed DJ, Martiny JBH. (2011). Patterns of fungal diversity and composition along a salinity gradient. ISME J 5: 379–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemergut DR, Anderson SP, Cleveland CC, Martin AP, Miller AE, Seimon A et al. (2007). Microbial community succession in an unvegetated, recently deglaciated soil. Microbiol Ecol 53: 110–122. [DOI] [PubMed] [Google Scholar]
- Ohtonen R, Fritze H, Pennanen T, Jumpponen A, Trappe J. (1999). Ecosystem properties and microbial community changes in primary succession on a glacier foreland. Oecologia 119: 239–246. [DOI] [PubMed] [Google Scholar]
- Olff H, de Leeuw J, Bakker JP, Platerink RJ, van Wijnen HJ. (1997). Vegetation succession and herbivory in a salt marsh: changes induced by sea level rise and silt deposition along an elevational gradient. J Ecol 85: 799–814. [Google Scholar]
- Pylro VS, Roesch LF, Ortega JM, do Amaral AM, Tótola MR, Hirsch PR et al. (2014). Brazilian microbiome project: revealing the unexplored microbial diversity - challenges and prospects. Microb Ecol 67: 237–241. [DOI] [PubMed] [Google Scholar]
- Richards TA, Jones MDM, Leonard G, Bass D. (2012). Marine fungi: their ecology and molecular diversity. Annu Rev Mar Sci 4: 495–522. [DOI] [PubMed] [Google Scholar]
- Salles JF, Poly F, Schmid B, Le Roux X. (2009). Community niche predicts the functioning of denitrifying bacterial assemblages. Ecology 90: 3324–3332. [DOI] [PubMed] [Google Scholar]
- Schrama M, Berg MP, Olff H. (2012). Ecosystem assembly rules: the interplay of green and brown webs during salt marsh succession. Ecology 93: 2353–2364. [DOI] [PubMed] [Google Scholar]
- Schrama M, Jouta J, Berg MP, Olff H. (2013). Food web assembly at the landscape scale: using stable isotopes to reveal changes in trophic structure during succession. Ecosystems 16: 627–638. [Google Scholar]
- Schoch CL, Seifert KA, Huhndorf S, Robert V, Spouge JL, Levesque CA et al. (2011). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proc Natl Acad Sci USA 109: 6241–6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DP, Peay KG. (2014). Sequence depth, not PCR replication, improves ecological inference from next generation DNA sequencing. PLoS One 9: e90234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedersoo L, Bahram M, Polme S, Koljalg U, Yorou NS, Wijesundera R et al. (2014). Global diversity and geography of soil fungi. Science 346: 1256688. [DOI] [PubMed] [Google Scholar]
- Torzilli AP, Sikaroodi M, Chalkley D, Gillevet PM. (2006). A comparison of fungal communities from four salt marsh plants using automated ribosomal intergenic spacer analysis (ARISA). Mycologia 98: 690–698. [DOI] [PubMed] [Google Scholar]
- Treseder KK, Lennon JT. (2015). Fungal traits that drive ecosystem dynamics on land. Microbiol Mol Biol Rev 79: 243–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbanová M, Šnajdr J, Baldrian P. (2015). Composition of fungal and bacterial communities in forest litter and soil is largely determined by dominant trees. Soil Biol Biochem 84: 53–64. [Google Scholar]
- Van Wijnen HJ, Bakker JP, de Vries Y. (1997). Twenty years of salt marsh succession on a Dutch coastal barrier island. J Coast Conservat 3: 9–18. [Google Scholar]
- Větrovský T, Baldrian P. (2013). Analysis of soil fungal communities by amplicon pyrosequencing: current approaches to data analysis and the introduction of the pipeline SEED. Biol Fert Soils 49: 1027–1037. [Google Scholar]
- Větrovský T, Kolařík M, Žifčáková L, Zelenka T, Baldrian P. (2015). The rpb2 gene represents a viable alternative molecular marker for the analysis of environmental fungal communities. Mol Ecol Resour e-pub ahead of print 19 August 2015; doi: 10.1111/1755-0998.12456. [DOI] [PubMed]
- Vilgalys R, Hester M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172: 4238–4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voříšková J, Brabcová V, Cajthaml T, Baldrian P. (2014). Seasonal dynamics of fungal communities in a temperate oak forest soil. New Phytol 201: 269–278. [DOI] [PubMed] [Google Scholar]
- Walker LR, Wardle DA. (2014). Plant succession as an integrator of contrasting ecological time scales. Trends Ecol Evol 29: 504–510. [DOI] [PubMed] [Google Scholar]
- Walker LR, Wardle DA, Bardgett RD, Clarkson BD. (2010). The use of chronosequences in studies of ecological succession and soil development. J Ecol 98: 725–736. [Google Scholar]
- White TJ, Bruns TD, Lee S, Taylor J. (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gefland DH, Sninsky JJ, White TJ (eds), PCR Protocols: A Guide to Method and Applications. Academic Press: San Diego, CA, USA, pp 315–322. [Google Scholar]
- Wilde P, Manal A, Stodden M, Sieverding E, Hildebrandt U, Bothe H. (2009). Biodiversity of arbuscular mycorrhizal fungi in roots and soils of two salt marshes. Environ Microbiol 11: 1548–1561. [DOI] [PubMed] [Google Scholar]
- Zhang NL, Xia JY, Yu XJ, Ma KP, Wan SQ. (2011). Soil microbial community changes and their linkages with ecosystem carbon exchange under asymmetrically diurnal warming. Soil Biol Biochem 43: 2053–2059. [Google Scholar]
- Zinger L, Shahnavaz B, Baptist F, Geremia RA, Choler P. (2009). Microbial diversity in alpine tundra soils correlates with snow cover dynamics. ISME J 3: 850–859. [DOI] [PubMed] [Google Scholar]
- Zinger L, Lejon DPH, Baptist F, Bouasria A, Aubert S, Geremia RA et al. (2011). Contrasting diversity patterns of crenarchaeal, bacterial and fungal soil communities in an alpine landscape. PLoS ONE 6: e19950. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.