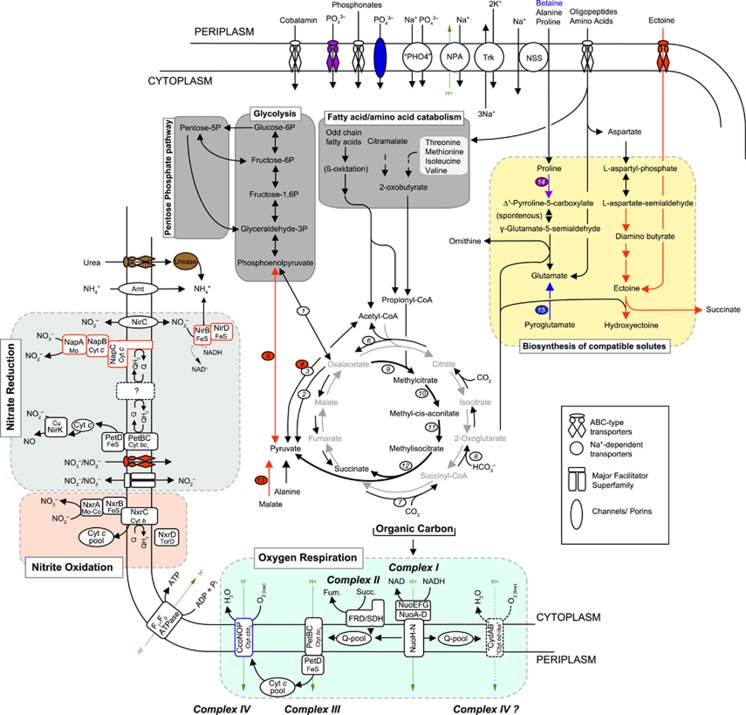
Figure 6.
Metabolic scheme of predicted pathways that are either unique to the pan-genome of Ca. Nitromaritima RS (in red), encoded only in the genome of Nitrospina gracilis (in blue), shared by both (in purple), present in all Nitrospina-like genomes (in black), or only in the other Ca. Nitromaritima SAGs (in brown). Dashed black arrows and a question mark symbol denote uncertain reactions, whereas grey and thick black arrows show the oxidative tricarboxylic acid cycle and the methylcitrate (anaplerotic) pathways, respectively. Important enzymes discussed in the main text are indicated with an oval shape: (1) phosphoenolpyruvate carboxykinase, (2) oxaloacetate decarboxylase, (3) pyruvate:ferredoxin oxidoreductase, (4) pyruvate dehydrogenase, (5) pyruvate-water dikinase, (6) ATP-dependent citrate lyase, (7) succinyl-CoA synthase, (8) oxoglutarate:ferredoxin oxidoreductase, (9) methylcitrate lyase, (10) methylcitrate dehydratase, (11) aconitase, (12) 2-methylisocitrate synthase, (13) NAD+-dependent malic enzyme, (14) proline dehydrogenase, (15) 5-oxoprolinase. NPA, sodium/proton antiporter; NSS, sodium:solute symporter; Trk, a potassium uptake system.