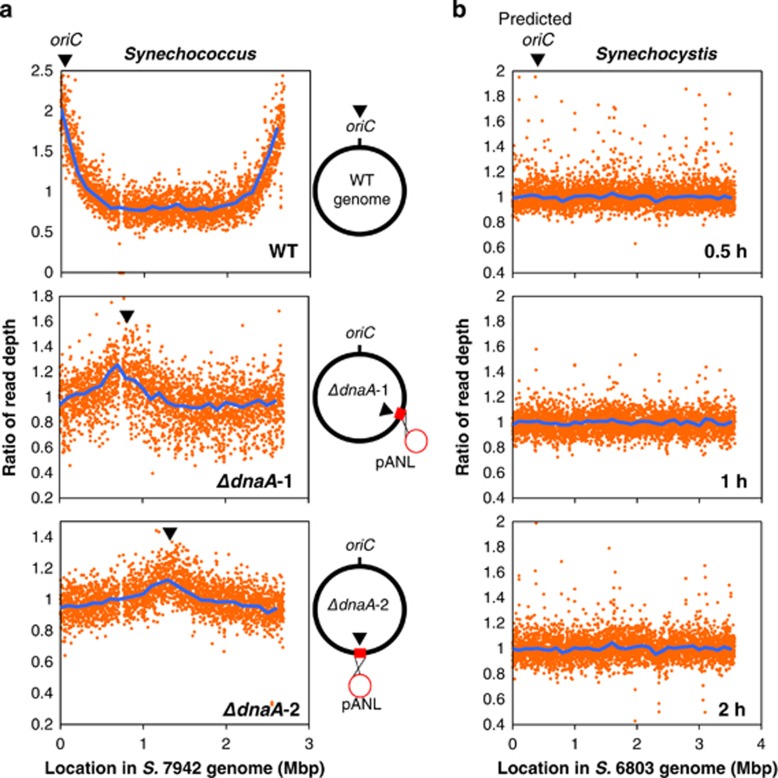
Figure 2.
Replication origin in S. elongatus and Synechocystis. Ratios of read depth at each genomic position analyzed by Repli-seq in WT and two dnaA disruptants of S. elongatus (a) and WT Synechocystis (b). (a) Synchronized cells were labeled with BrdU for 4 h; libraries of BrdU-labeled DNA were analyzed by next-generation sequencing and mapped using the S. elongatus (S. 7942) genome as a reference. Genome maps are shown to the right of the depth plot. Inverted triangles denote replication initiation points. (b) Synchronized Synechocystis cultures were labeled with BrdU for 0.5, 1 and 2 h. Sequence reads were mapped using the Synechocystis (S. 6803) genome as a reference.