FIGURE 4.
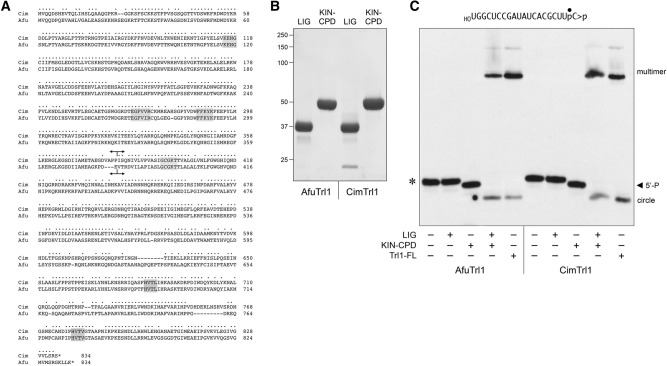
Separate healing and sealing domains of AfuTrl1 and CimTrl1. (A) Alignment of the primary structures of AfuTrl1 and CimTrl1. Positions of side chain identity/similarity are indicated by (•). Gaps in the alignment are denoted by dashes. Signature active site motifs of the ligase, kinase, and CPDase enzymes are shaded in gray boxes. Forward and reverse arrowheads indicate the boundaries of the N-terminal LIG and C-terminal KIN-CPD domain constructs, respectively. (B) Aliquots (5 µg) of the recombinant AfuTrl1 and CimTrl1 LIG and KIN-CPD preparations were analyzed by SDS-PAGE. The Coomassie blue-stained gel is shown. The positions and sizes (kDa) of marker proteins are indicated on the left. (C) Reaction mixtures (20 µL) containing 50 mM Tris–HCl (pH 7.5), 2 mM DTT, 10 mM MgCl2, 100 µM ATP, 100 µM GTP, 20 nM 20-mer HORNA>p (depicted at the top), and 50 nM LIG, KIN-CPD, or full-length (FL) Trl1 proteins (as specified by + below the lanes) were incubated for 5 min at 22°C. The products were analyzed by urea–PAGE. An autoradiograph of the gel is shown. The positions and identities of the HORNA>p substrate (*), 5′-phosphorylated kinase product (5′-P), and ligation products (circle and multimers) are indicated.