FIGURE 3.
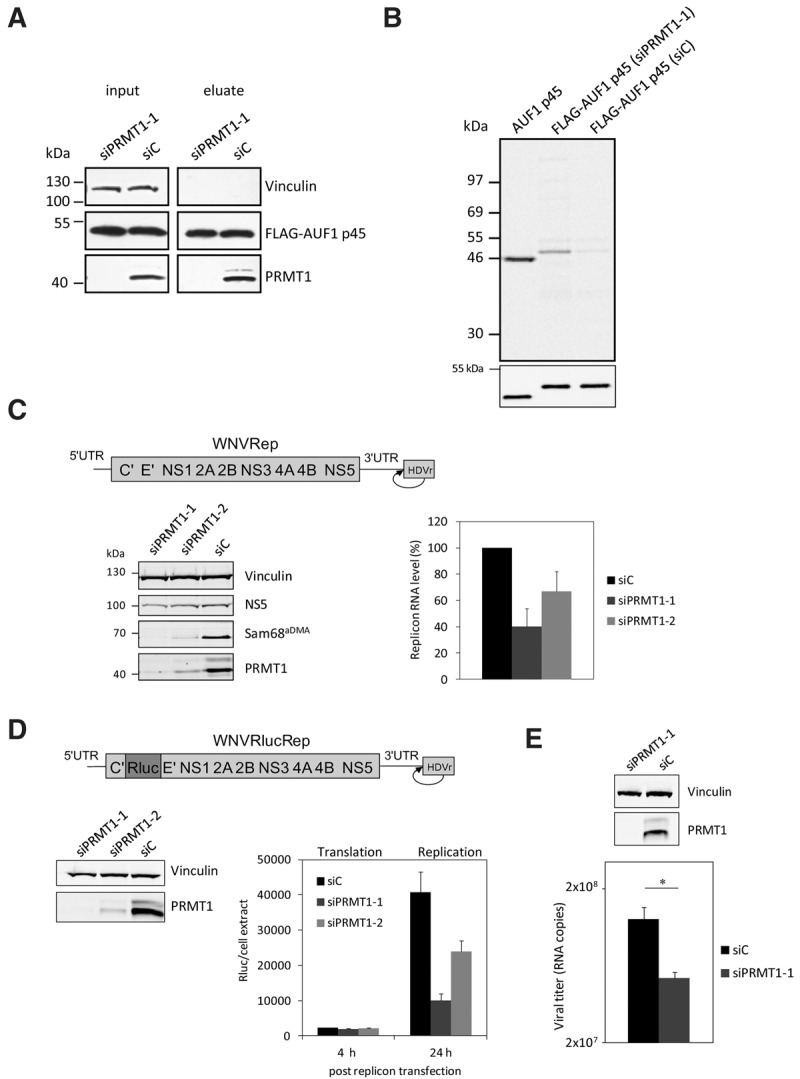
RNAi-mediated depletion of PRMT1 in Huh7 cells inhibits the methylation of FLAG–AUF1 p45 and WNV replication. (A) AUF1 p45 was expressed as a FLAG fusion protein in Huh7 cells and immune-affinity purified from extracts of cells that were earlier treated with a control siRNA (siC) or with an siRNA targeting the PRMT1 encoding mRNA (siPRMT1-1). The amounts of expressed FLAG–AUF1 p45 were determined by Western blot with anti-FLAG antibody comparing similar protein amounts of the original extracts (input) and of the eluate, respectively. The efficient depletion of PRMT1 by siPRMT1-1 was verified at 120 h post-transfection by Western blot with an anti-PRMT1 antibody. (B) In vitro methylation assay with PRMT1 and different preparations of AUF1 p45. Applied were 5 pmol of nonmethylated AUF1 p45 (expressed and purified from E. coli) and comparable amounts of FLAG–AUF1 p45 that was expressed in and purified from Huh7 cells (transfected with either siC or siPRMT1-1). The different AUF1 p45 preparations were methylated in vitro by 5 pmol of purified PRMT1 v1 using 400 pmol [S-14C] adenosylmethionine as a substrate. The samples were taken after 2 h and the methylation state analyzed by SDS–PAGE and phosphorimaging. (Lower panel) Western blot with anti-AUF1 antibody demonstrating the use of equal amounts of protein in the in vitro methylation assay. One representative experiment is shown. Note that the E. coli and Huh7-derived proteins show a slightly different migration behavior in the SDS–PAGE; this was explained by the FLAG-affinity tag that is present in the Huh7-expressed protein. (C) Scheme of the organization of the applied WNV replicon; the ORF is represented as a bar, the UTRs as single lines. The authentic 3′-end of the viral RNA is generated by the activity of a hepatitis delta ribozyme (HDVr) that was fused to the 3′ UTR (as indicated). Huh7 cells were transfected with control siRNA (siC) or siRNAs targeting the PRMT1 encoding mRNA (siPRMT1-1 and siPRMT1-2), respectively. Seventy-two hours later, all cells were transfected with the WNV replicon RNA. At 24 h post-transfection of the replicon, the protein levels of PRMT1, the viral NS5, Vinculin and of the methylated Sam68 were analyzed by Western blotting using appropriate antibodies (left panel; representative experiment shown). The RNA levels of the WNV replicon were quantified by real-time PCR at 24 h post-transfection (right panel). Results of three different experiments are shown; error bars reflect standard deviations. (D, top) Schematic representation of the applied WNVRluc replicon; the integrated Rluc ORF is indicated by different shading. Huh7 cells were treated with a control siRNA (siC) and siRNAs (siPRMT1-1 or siPRMT1-2) targeting PRMT1, respectively. The cytoplasmic depletion of PRMT1 120 h post siRNA transfection was confirmed by Western blotting (left), and the cells were transfected with the WNVRluc replicon RNA. At 4 h and 24 h post-transfection, the Renilla luciferase activity was measured (right). Results from three different experiments are shown; error bars reflect standard deviations. (E) Huh7 cells were treated with a control siRNA (siC) and an siRNA targeting PRMT1 (siPRMT1-1). After 120 h, the cytoplasmic depletion of PRMT1 was confirmed by Western blotting (top), and the cells were infected with WNV strain 3356. At 24 h post-infection, culture fluids were assayed for virus titers by qRT-PCR (see Materials and Methods). One representative experiment (performed in triplicate) is shown; (*) P ≤ 0.05.
