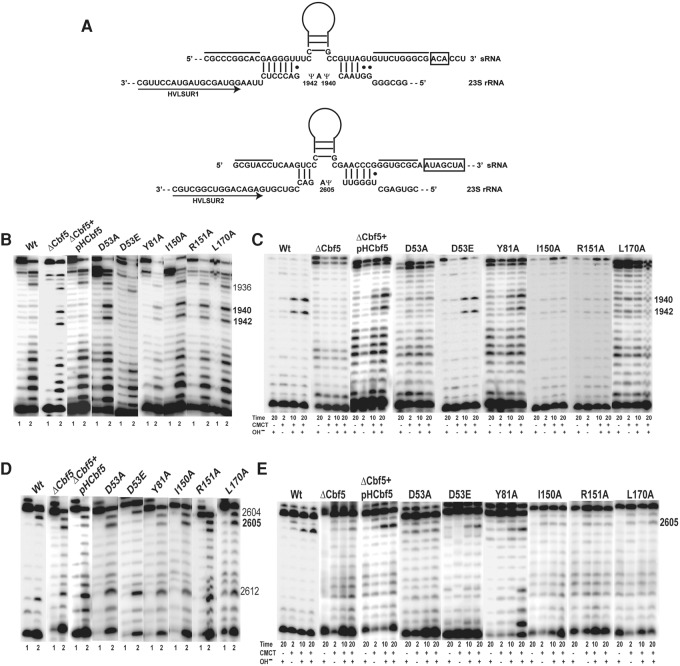
FIGURE 3.
Ala substitutions of HvCbf5 residues conserved across all Ψ synthases abolish or reduce Cbf5-mediated Ψ formation at the three positions of 23S rRNA. (A) Interactions between the two H/ACA motifs of the sRNA and their 23S rRNA target sequences are shown. The two lines above the sRNA indicate the sequences that pair to form the lower stem of the H/ACA RNA. H and ACA sequences are boxed. Regions of sRNAs above the Ψ pocket are illustrated as stem–loops. Positions of the primers (arrows) HVLSUR1 and HVLSUR2 relative to rRNA sequences are shown. Positions of Ψ’s (1940, 1942, and 2605) in the rRNA sequence are indicated. (B) U-specific analyses to determine the modification status of U1940 and U1942 of 23S rRNA were done using primer HVLSUR1 (see panel A) and total RNA of Δcbf5 strain transformed with different mutant pHCbf5 plasmids (marked above each panel). Lanes 1 and 2: Primer extensions on untreated RNA and on RNA following U-specific reactions, respectively. A dark band at a position in lane 2, but not in lane 1 indicates an unmodified U. Positions of certain U's in 23S rRNA are indicated on the side. In WT cells, U1940 and U1942 are converted to Ψ, and U1936 (used as indicator for the positions) remains unmodified. (C) The primer and total RNA used in B are also used for CMCT-primer extension analyses. Total RNAs were either untreated (−) or treated with CMCT (+) for the indicated time (in minutes), followed by alkali (OH−) treatment (+) or no treatment (−). Positions of Cbf5-mediated modifications are indicated on the side. A dark band in CMCT followed by alkali, in 10 min and 20 min lanes, indicates Ψ. (D,E) Analyses similar to those in B and C, respectively, using primer HVLSUR2 (see panel A), were done to determine the modification status of U2605 of 23S rRNA. Unmodified U2604 and U2612 served as indicators for positions in D.