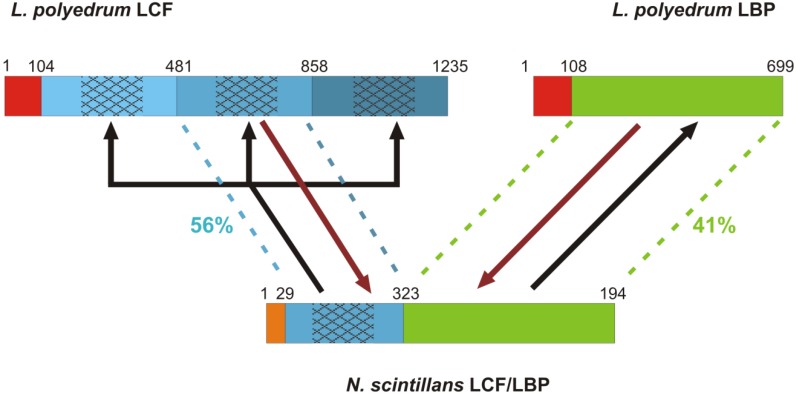
Figure 2.
A schematic representation of the different LCF and LBP structures labeled with amino acid positions and the two scenarios for their evolution [30,32]. Lingulodinium polyedrum LCF represents the photosynthetic dinoflagellates in general [30]. The three non-identical repeat domains of L. polyedrum LCF (different shades of blue) are each 377 amino acids long with 150 amino acids in the central regions that encode the catalytic sites (patterned) and retain higher sequence similarity. The single LCF domain of N. scintillans is most similar to the second domain of L. polyedrum LCF. The LBP domain in N. scintillans is structurally equivalent to that in L. polyedrum, both consisting of four repeat domains. In both organisms, LCF is preceded by an N-terminal gene region which is similar in the LCF and LBP of L. polyedrum (red) but not in N. scintillans (orange). Two potential scenarios of how these gene structures arose are shown. In the gene fission scenario (black line), the N. scintillans gene split and the LCF domain underwent successive duplications. In the gene fusion scenario (brown lines), the second domain of LCF was excised (e.g., by splicing) and fused with LBP in N. scintillans.