Figure 1. Involvement of miR-16 in Taxol chemosensitivity in breast cancer cells.
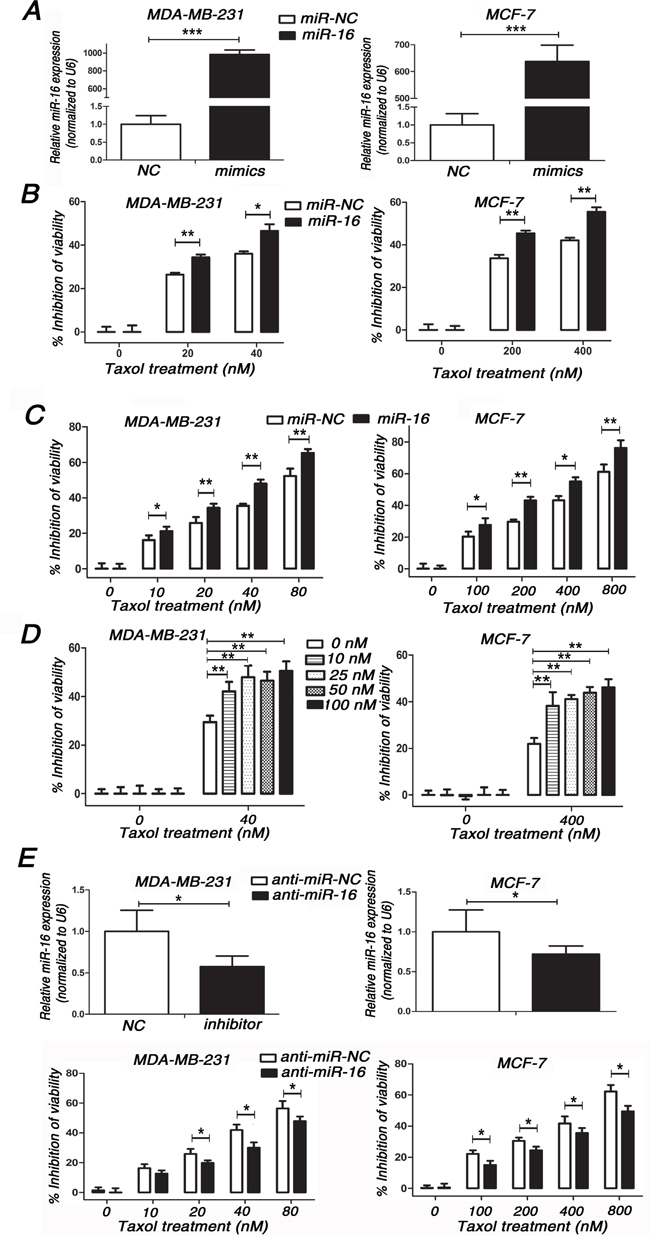
A. miR-16 levels were detected in MDA-MB-231 and MCF-7 cells transfected with 50 nM miR-16 mimics or 50 nM miR-NC by qRT-PCR. B. MDA-MB-231 and MCF-7 cells transfected with 50 nM miR-16 mimics or 50 nM miR-NC were seeded into 96-well plates and treated with 0, 20, 40 nM (MDA-MB-231) or 0, 200, 400 nM (MCF-7) Taxol for 48 h. The cell viabilities were detected by MTT assays. C. MDA-MB-231 and MCF-7 cells transfected with 50 nM miR-16 mimics or 50 nM miR-NC were seeded into 96-well plates and treated with 0, 10, 20, 40, 80 nM (MDA-MB-231) or 0, 100, 200, 400, 800 nM (MCF-7) Taxol for 48 h. The cell viabilities were detected by MTT assays. D. MDA-MB-231 and MCF-7 cells transfected with 0, 10, 25, 50, 100 nM miR-16 mimics or equal dose of miR-NC were seeded into 96-well plates and treated with 0, 40 nM (MDA-MB-231) or 0, 400 nM (MCF-7) Taxol for 48 h. The cell viabilities were detected by MTT assays. E. MDA-MB-231 and MCF-7 cells transfected with 100 nM miR-16 inhibitor or 100 nM anti-miR-NC (top) were seeded into 96-well plates and treated with 0, 10, 20, 40, 80 nM (MDA-MB-231) or 0, 100, 200, 400, 800 nM (MCF-7) Taxol for 48 h. The cell viabilities were detected using MTT assays. Data are presented as the percentage of viability inhibition measured in untreated cells. Columns, means of three independent experiments; bars, SE. *, p<0.05, **, p<0.01, ***, p<0.001.
