Figure 5. DEK is recruited to the DRE and HRE sites of VEGF promoter and promotes the recruitment of HIF-1α and p300.
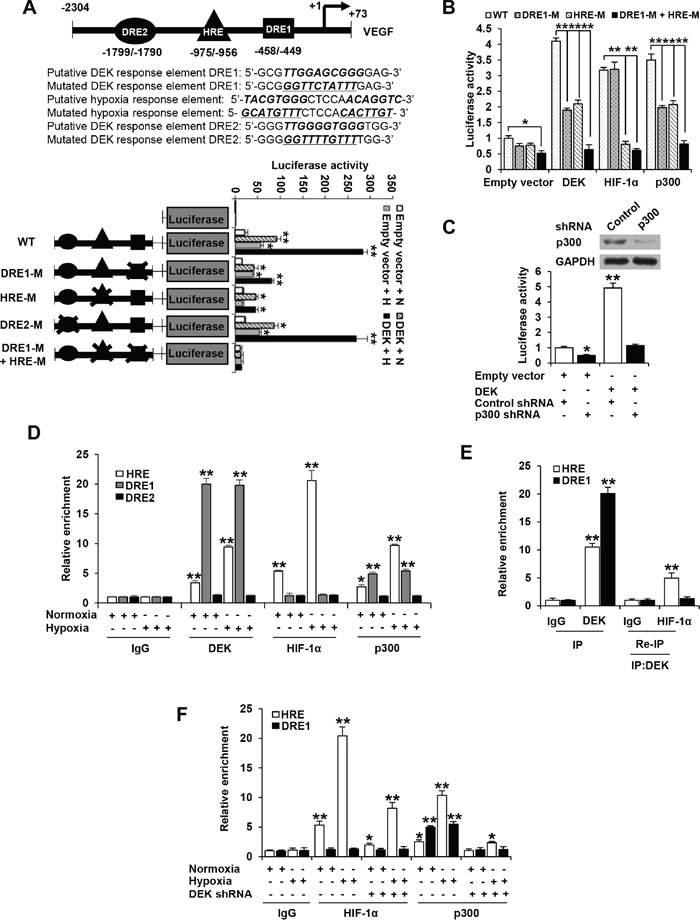
A. Luciferase activity of different VEGF promoter constructs in MCF-7 cells transfected with DEK or empty vector. At 24 h post-transfection, cells were exposed to either normoxic (N) or hypoxic (H) conditions for another 24 h. The arrow indicates the position of the transcriptional start site. DRE1 and DRE2 are putative DEK binding sites, and HRE is HIF-1α binding site. The “X” shows the mutated putative DEK-binding sites or HIF-1 binding site. B. Luciferase reporter assays were performed in MCF-7 cells cotransfected with indicated VEGF promoter constructs from A and DEK, HIF-1α or p300. The transfected cells were harvested for measurement of luciferase activities of different promoter constructs. C. Luciferase reporter assays were performed in MCF-7 cells cotransfected with the VEGF-Luc reporter and p300 shRNA. The transfected cells were harvested for measurement of VEGF-Luc activity. Representative immunoblot indicates the expression of p300. Data shown are mean ± SD of triplicate measurements and have been repeated 3 times with similar results. *P < 0.05, **P < 0.01 versus empty vector with corresponding promoter reporter (A-C). D. ChIP analysis of the occupancy of DEK, HIF-1α and p300 on DRE1, DRE2 or HRE of VEGF promoter in MCF-7 cells under normoxic or hypoxic conditions for 24 h. E. Re-ChIP analysis of the occupancy of DEK and HIF-1α on DRE1 and HRE of VEGF promoter in MCF-7 cells under hypoxic condition for 24 h. DEK formed a complex with HIF-1α on the HRE site but not on the DRE1 site. F. ChIP analysis were performed to detect the occupancy of HIF-1α and p300 on DRE1 and HRE of VEGF promoter in MCF-7 cells stably infected with lentivirus carrying DEK shRNA or control shRNA under normoxic or hypoxic conditions for 24 h. All values shown are mean ± SD of triplicate measurements and have been repeated 3 times with similar results (D-F). *P < 0.05, **P < 0.01 versus corresponding control (D-F).
