Figure 4.
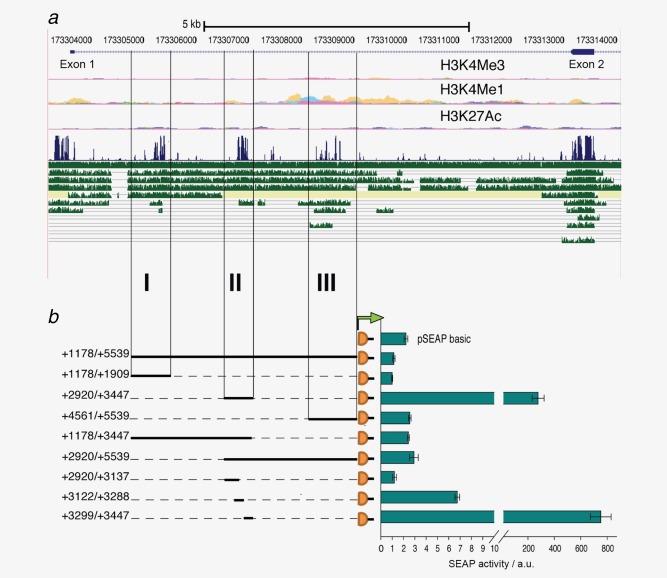
Analysis of the evolutionary conserved regions within the first intron. (a) UCSC browser display of the 5′ region of the tenascin‐W gene with absolute coordinates of chromosome 1 indicated above the RefSeq genes showing Exon 1 and 2 separated by a large Intron. Chromatin signatures shown below (H3K4Me3, H3K4Me1 and H3K27Ac) overlap with the intronic modules. Below the Vertebrate Multiz Alignment & Conservation track in blue, tracks in green show from top to bottom comparisons with rhesus, mouse, dog, horse, armadillo, opossum, platypus, lizard, chicken, X_tropicalis and stickleback orthologs. (b) To assess potential transcriptional activities, each conserved region I, II and III, combinations thereof and truncated versions as indicated were cloned upstream of the SEAP reporter gene. Transient transfections of the constructs in HT1080 cells were analyzed for SEAP activity shown in arbitrary units (a.u.) normalized to a co‐transfected secreted luciferase plasmid. The experiment was performed in triplicates and repeated three times (error bars = SEM). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
