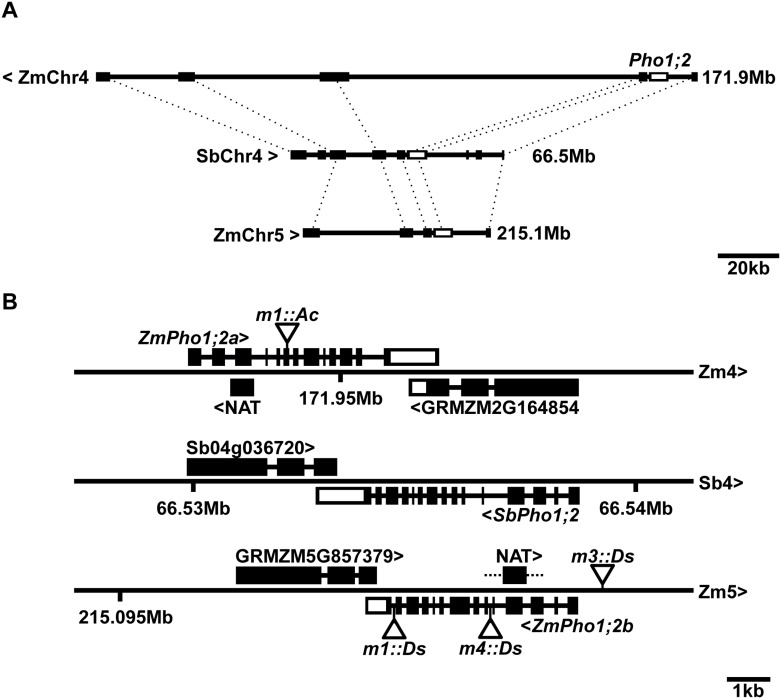
Fig 2.
Microsynteny among Pho1;2 loci A) Annotated genes (filled boxes) in the region ofSbPho1;2 on Sorghum chromosome 4 (SbChr4) and candidate orthologs on maize (B73) chromosomes 4 (ZmChr4) and 5 (ZmChr5). Orthologous genes are connected by dashed lines. Pho1;2 genes on the three chromosomes shown in unfilled boxes. Regions shown to scale, the right hand position indicated. SbChr4 and ZmChr5 run left to right, ZmChr4 right to left. B) Pho1;2 gene models on ZmChr4, SbChr4 and ZmChr5. Exons are shown as boxes, coding regions filled, UTR unfilled. Putative anti-sense transcripts associated with ZmPho1;2a and ZmPho1;2b shown as filled boxes (NAT). Angle brackets indicate the direction of transcription. The maize syntenic paralog pair GRMZM2G164854/GRMZM5G853379 and their sorghum ortholog are also shown. Triangles indicate the position of Activator and Dissociation insertion. Refer to Table 4 for full description alleles. Shown to scale.