Fig. 4. aPBPs can function independently from the cytoskeletal machinery.
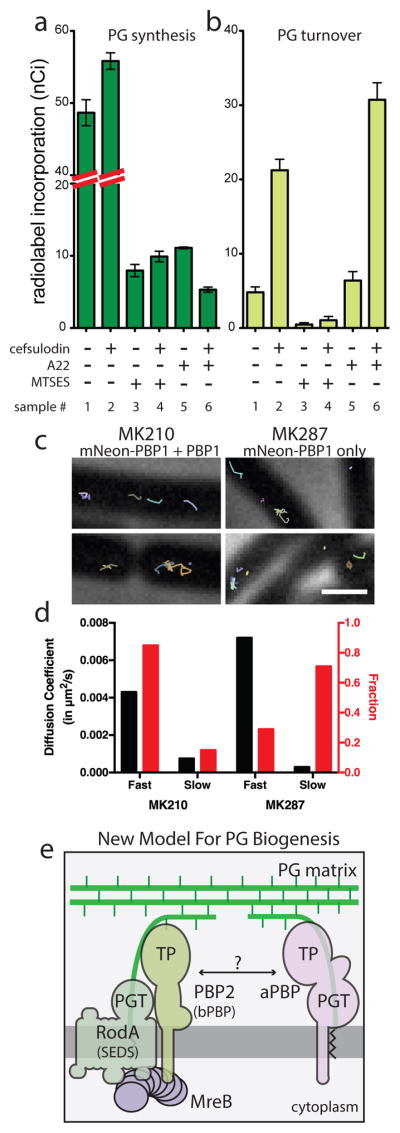
A–B. PG matrix assembly and turnover were measured as in Figure 2 using strain HC533(attλC739) [ΔlysA ΔampD ΔponA ΔpbpC ΔmtgA MSponB (Ptac::sulA)]. Cefsulodin was used at 100 μg/ml. Results are the average of three independent experiments with the error bars representing the SEM. C. Tracks of mNeon-PBP1 expressed as (right) the only copy or (left) in addition to native untagged protein in B. subtilis. Each continuously tracked particle is highlighted with a different color trajectory. Note that although no MreB-like directional motion was observed, particles occasionally travel rapidly in one direction for a few frames as expected for membrane diffusion. D. Graph showing diffusion constants, and fraction of particles tracked in each diffusion state as determined by CDF analysis. Microscopy results are representative of at least two independent experiments. E. Schematic view of a new model for PG biogenesis involving two different classes of PG polymerases working semi-autonomously. SEDS PGTs and partner bPBPs perform PG polymerization and crosslinking in the context of the Rod system and divisome (not shown) while aPBPs function outside of these complexes. Collaboration between the synthases likely occurs but the mechanism remains to be defined.
