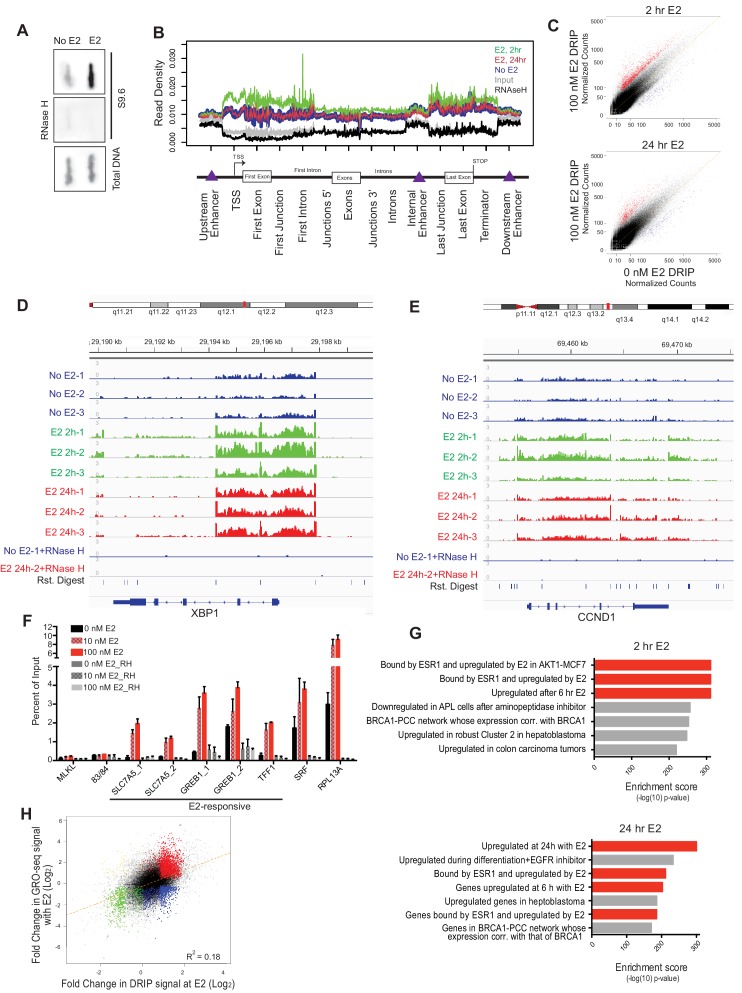
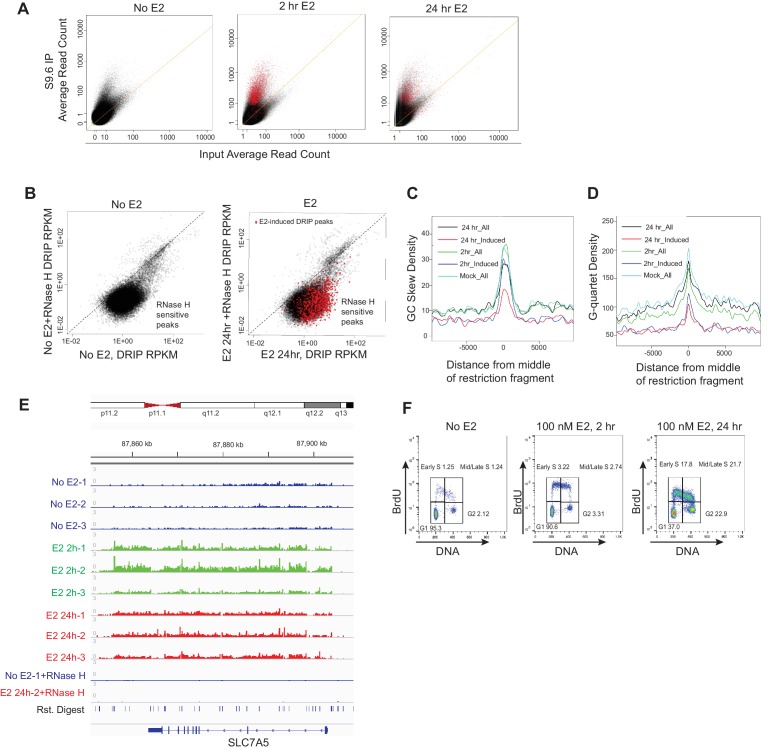
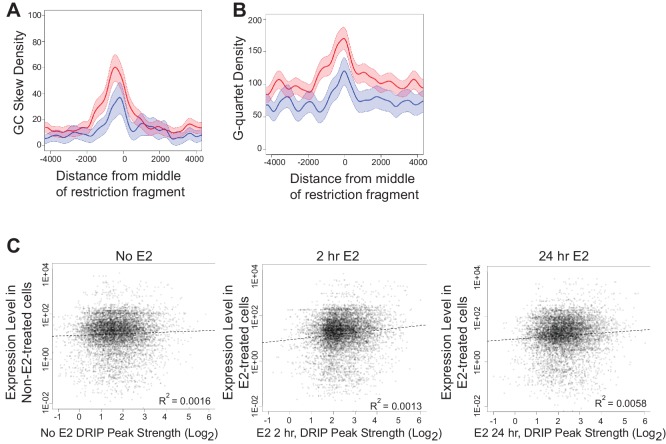
Figure 2. Estrogen induces robust R-loop formation at E2-responsive genes.
(A) Slot blot to detect global RNA-DNA hybrids with S9.6 antibody in MCF7 cells treated with 0 or 100 nM E2 for 24 hr. Total denatured DNA is stained with a single-strand DNA antibody. RNase H was added as indicated. (B) Meta-gene analysis for indicated DRIP signal over indicated genomic features. Data are shown for DRIP-seq biological replicate # 3. An enrichment is seen in all data sets but relative read density at these sites varies between replicates. (C) DRIP-seq read counts normalized for total mapped reads from DRIP in 0 nM E2 conditions vs DRIP from cells treated with 100 nm E2 for 2 hr (top) or 24 hr (bottom). Graphs are from 3 biological experiments. Black dots indicate DRIP peaks and red dots indicate induced DRIP peaks relative to 0 nM E2. (D) Integrated Genome Viewer (IGV) display of DRIP-seq enrichment at XBP1. Scale = million mapped reads. RNase H was performed prior to DRIP-seq on one replicate each from 0 nM E2 and 100 nM E2 24 hr. Independent replicates are shown as 1–3. (E) IGV display of CCND1 (Cyclin D1), as described in (D). (F) DRIP-qPCR validation. Cells were treated with 0, 10, or 100 nM E2 for 24 hr and harvested for DRIP. MLKL and 83/84 are negative controls. Error bars represent S.E.M. of 2 biological experiments. RNase H treatment was performed for 24 hr prior to DRIP-qPCR where indicated. (G) Functional signatures by GREAT of E2-induced DRIP peaks found to be differentially induced in 100 nM E2, 2 hr (top) or 100 nM E2 24 hr (bottom) than in 0 nM E2 treated cells. The 7 highest enrichment scores are shown, with red highlighting E2-associated signatures. (H) Fold change in DRIP signal after 2 hr of 100 nM E2 relative to 0 nM E2 (x-axis) vs. fold change in GRO-seq signal after 160 min of 100 nM E2 relative to 0 nM E2 (y-axis). E2-induced DRIP peaks that show a positive (red) or negative (blue) fold change in GRO-seq upon E2 stimulation are highlighted. Negative changes in DRIP upon E2 that correspond to a positive (yellow) or negative (green) fold change in GRO-seq are also shown. GRO-seq data from (Hah et al., 2011).
DOI: http://dx.doi.org/10.7554/eLife.17548.010