Abstract
Moonmilk speleothems of limestone caves host a rich microbiome, among which Actinobacteria represent one of the most abundant phyla. Ancient medical texts reported that moonmilk had therapeutical properties, thereby suggesting that its filamentous endemic actinobacterial population might be a source of natural products useful in human treatment. In this work, a screening approach was undertaken in order to isolate cultivable Actinobacteria from moonmilk of the Grotte des Collemboles in Belgium, to evaluate their taxonomic profile, and to assess their potential in biosynthesis of antimicrobials. Phylogenetic analysis revealed that all 78 isolates were exclusively affiliated to the genus Streptomyces and clustered into 31 distinct phylotypes displaying various pigmentation patterns and morphological features. Phylotype representatives were tested for antibacterial and antifungal activities and their genomes were mined for secondary metabolite biosynthetic genes coding for non-ribosomal peptide synthetases (NRPSs), and polyketide synthases (PKS). The moonmilk Streptomyces collection was found to display strong inhibitory activities against a wide range of reference organisms, as 94, 71, and 94% of the isolates inhibited or impaired the growth of Gram-positive, Gram-negative bacteria, and fungi, respectively. Interestingly, 90% of the cave strains induced strong growth suppression against the multi-drug resistant Rasamsonia argillacea, a causative agent of invasive mycosis in cystic fibrosis and chronic granulomatous diseases. No correlation was observed between the global antimicrobial activity of an individual strain and the number of NRPS and PKS genes predicted in its genome, suggesting that approaches for awakening cryptic metabolites biosynthesis should be applied to isolates with no antimicrobial phenotype. Overall, our work supports the common belief that moonmilk might effectively treat various infectious diseases thanks to the presence of a highly diverse population of prolific antimicrobial producing Streptomyces, and thus may indeed constitute a promising reservoir of potentially novel active natural compounds.
Keywords: geomicrobiology, secondary metabolism, MLSA phylogeny, cryptic antibiotics, genome mining
Introduction
Members of the phylum Actinobacteria can be found in all kinds of extreme environments (Saiz-Jimenez, 1999; Bull, 2010; Prieto-Davó et al., 2013; Zhang et al., 2014; Mohammadipanah and Wink, 2015; Shivlata and Satyanarayana, 2015). Their successful survival in severe conditions suggests broad adaptive abilities that might be directly related to their very diverse and specialized (secondary) metabolism. Natural small molecules, collectively termed the parvome (Davies and Ryan, 2012), apart from having essential ecological functions, possess a wide range of bioactivities, which are applicable for agro-industrial purposes and human/animal therapy (Hopwood, 2007). Since the metabolome of soil-dwelling Actinobacteria, especially of the members of the genus Streptomyces, has been widely exploited, leading to the multiple re-isolation of already known bioactive compounds, the attention has been refocused toward unexplored and extreme environments, which can potentially be a source of novel species and consequently of novel molecules of interest (Cheeptham et al., 2013; Claverías et al., 2015; Mohammadipanah and Wink, 2015; Liao et al., 2016).
The geological isolation of caves from surface processes makes them a unique niche, not only to study microbial interactions and adaptations to extreme oligotrophy, but also to screen for potentially novel bioactive compounds. Although, the most common Actinobacteria reported from caves belong to Pseudonocardiaceae and Nocardiaceae families (Stomeo et al., 2008; Porca et al., 2012; Quintana et al., 2013; Riquelme et al., 2015), many investigations have identified cultivable members of the genus Streptomyces, which are the most prolific antimicrobial producers (Cañaveras et al., 1999; Groth et al., 1999; Cheeptham et al., 2013; Nimaichand et al., 2015; Axenov-Gribanov et al., 2016). The presence of Actinobacteria as dominant members of microbial ecosystems in caves is puzzling, as these bacteria, particularly Streptomyces are often presented as major protagonists in the recycling of the residual plant biomass in nutrient-rich soil environments (Hodgson, 2000). Nonetheless, >99% of allochtonous carbon entering caves, primarily with drip water, contains soil-derived dissolved organic carbon in the form of partially degraded plant and fungal polymers, which can be catabolized by the enzymatic arsenals of Streptomyces (Saiz-Jimenez and Hermosin, 1999; Simon et al., 2007; Barton, 2015). Additionally, highly prolific and diversified secondary metabolism could be a driving force in the dominance of these species in oligotrophic environments, through which they could shape microbiomes thanks to their specialized metabolites, such as metal-chelators for acquiring trace metals, and antibiotics to prevent nutrient-exclusion by competitive species (Bhullar et al., 2012). Antibiotics do not exclusively prevent microbial growth, but are also known to act as inter-/intraspecies communication (Sengupta et al., 2013), and as cues triggering adaptations, such as motility or biofilm formation (Linares et al., 2006), or can be used as alternative carbon and/or nitrogen sources (Dantas et al., 2008). Consequently, small bioactive molecules expressed under nutrient-starved conditions, could be used as weapons, as signals, or as nutrient sources.
Moonmilk is a comparatively rare speleothem in cave environments, where it forms as a thick calcite paste (similar in consistency to toothpaste) up to 10s-of-centimeters in thickness in passageways that receive significant airflow (Hill and Forti, 1997; Borsato et al., 2000). The exact process of moonmilk formation remains in debate; however, the consistency, the high abundance of filamentous bacterial cells and members of the Actinobacteria has led researchers to suggest a biogenic origin (Cañaveras et al., 2006; Rooney et al., 2010; Portillo and Gonzalez, 2011). Despite its relative scarcity in caves, moonmilk has had long scientific interest due to its historical use as a medical treatment. Interestingly, moonmilk was used in human and animal therapy since the Middle Ages (Reinbacher, 1994). Its curative properties could be associated with the presence of the numerous filamentous Actinobacteria, particularly Streptomyces, presumably producing bioactive molecules. Indeed, isolation of Streptomyces with antimicrobial and antifungal activities has been reported for moonmilk from the Bolshaya Oreshnaya Cave in Siberia (Axenov-Gribanov et al., 2016). The identification of potential novel compounds with broad-spectrum activities from Streptomyces found in this karstic secondary deposit supports the idea of moonmilk being a great target for bioactivity screening, although to date very few studies have examined cultivable moonmilk microbiome and the diversity of its metabolome as a possible reservoir of novel compounds. The fact that caves are unique and still highly under-explored environment increase the chances of finding novel organisms and consequently novel bioactive compounds that might be useful in the context of the global health problem of antibiotic resistance (Bush et al., 2011; Laxminarayan et al., 2013; Berendonk et al., 2015; Frère and Rigali, 2016).
In this work, we report the isolation and phylogenetic analysis of a collection of novel Streptomyces isolates from moonmilk deposits, and assess their potential as producers of compounds with antimicrobial and antifungal properties through in vitro screening and genome mining approaches.
Materials and Methods
Site Description and Sampling
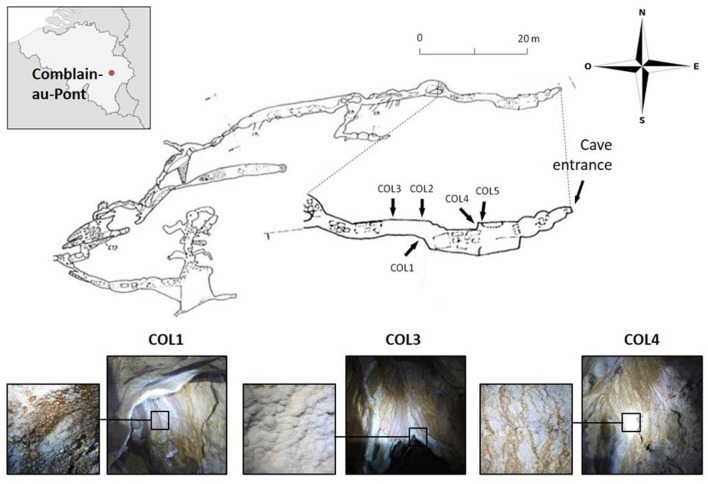
The cave called Grotte des Collemboles (Springtails’ Cave) located in Comblain-au-Pont in Belgium is a shallow (<20 m), ∼70 m long fissure cave, formed in the upper Viséan limestone (Figure 1), with an average annual temperature of ∼11.5°C. Due to cave protection policies and location on private property, specific location details and access information is available to other researchers upon request. Within the cave, white to brown-orange (presumably from iron-oxide precipitates) moonmilk deposits are found on the walls within the first 20 m of the cave in the first narrow chamber located at the entrance of the cave as well as in the narrow passages leading deeper into the cave (Figure 1). Samples used in this work were aseptically collected in January 2012 from three moonmilk deposits (Figure 1). Soft moonmilk speleothem was scratched with sterile scalpels from the wall in the first chamber, adjacent to the cave entrance (COL4) and the walls in a narrow passage after the first chamber (COL1, COL3; Figure 1). Samples were collected into falcon tubes, transferred to the laboratory, freeze-dried on a VirTis Benchtop SLC Lyophilizer (SP Scientific, Warminster, PA, USA) and stored at -20°C.
FIGURE 1.
Cave map of the Grotte des Collemboles and visualization of the moonmilk deposit sampling points. Location of the Grotte des Collemboles (Springtails’ Cave) in Comblain-au-Pont (Liège, Belgium) and cave map with general view and close up of the moonmilk deposits from the different collection points (COL).
Isolation of Cultivable Actinobacterial Species
Selective isolation of Streptomyces species from moonmilk was carried out by a serial dilution method as described previously (Maciejewska et al., 2015). 250 mg of lyophilized moonmilk sample from each collection point was suspended in 0.25X strength Ringer’s solution supplemented with 0.001% Tween 80. Resulting moonmilk suspensions were serially diluted in PBS and inoculated in duplicates on ISP media (Shirling and Gottlieb, 1966), starch nitrate (SN) medium (Gauze’s medium No.1; Waksman and Lechevalier, 1961), B-4 agar (Boquet et al., 1973), and minimal medium (Kieser et al., 2000) with 1% chitin (MMch). Isolation media were supplemented with nalidixic acid (75 μg/ml) and nystatin (50 μg/ml) to suppress the growth of Gram-negative bacteria and fungi, respectively. After 1 month of incubation at 17°C, colony forming units (CFUs) were enumerated and 129 isolates were selected. After two rounds of subcultivation, 78 isolates were recovered as purified strains and subsequently preserved both on ISP2 slopes at 4°C and as 25% glycerol mycelium stock at -20°C.
DNA Extraction, Genome Sequencing, and Gene Selection from Moonmilk-Derived Isolates
In order to screen for the genes of interest, which would enable to identify moonmilk derived isolates, to perform phylogenetic analysis, as well as to investigate strains antimicrobial properties, de novo genome sequencing was carried out. DNA from purified strains was extracted with GenElute Bacterial Genomic DNA Kit (Sigma-Aldrich, St. Louis, MO, USA) according to manufacturer’s instructions from liquid LB (Luria-Bertani; Difco, BD, Franklin Lakes, NJ, USA) cultures incubated at 28°C. The genomic libraries of moonmilk isolates were constructed using Nextera XT kit (Illumina, Inc., San Diego, CA, USA). Library concentrations and mean fragment lengths were measured by Qubit fluorometer (Invitrogen, Grand Island, NY, USA) and Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), respectively. De novo sequencing with 2 × 250 bp and 2 × 300 bp reads configuration was carried out on the Illumina MiSeq platform (Illumina, Inc., San Diego, CA, USA) at the Luxembourg Institute of Science and Technology. Complete genomes were assembled from raw sequence data with SPAdes v.3.6.2 (Bankevich et al., 2012) using the “careful” option, and the quality of the assemblies was subsequently assessed with QUAST v2.3 (Gurevich et al., 2013).
To infer the evolutionary relationships between moonmilk strains and their closest relatives, as well as between cave isolates themselves, 16S rRNA-based phylogeny was combined with multilocus sequence analysis (MLSA). For this purpose, along with the 16S rRNA gene, five additional housekeeping genes were selected, namely atpD, gyrB, recA, rpoB, and trpB (Han et al., 2012). In order to identify these genes within moonmilk genomes, the corresponding nucleotide sequences (16S rRNA) and protein translations (atpD, gyrB, recA, rpoB, and trpB) were retrieved from the NCBI web portal for three reference strains: Streptomyces peucetius AS 4.1799, Streptomyces pristinaespiralis ATCC 25486 and Streptomyces venezuelae ATCC 10712 (Supplementary Table S1). Core alignments were built using MAFFT v7.273 (Katoh and Standley, 2013) with default parameters, then enriched in the corresponding sequences from moonmilk genomes using the software “42” (D. Baurain, to be published elsewhere), which mines genomic contigs for orthologous genes and aligns the (translated) identified sequences on their closest relatives. Enriched alignments were then refined by hand using the ED program of the MUST software package (Philippe, 1993). Finally, protein sequences were turned into nucleotide sequences using the software “1331” (D. Baurain, to be published elsewhere), which uses a protein alignment as a guide to generate the corresponding nucleotide alignment from genomic contigs. The sequences of the five protein coding housekeeping genes for all the moonmilk isolates were deposited in GenBank and the corresponding accession numbers are given in Supplementary Table S2, while Table 1 and Supplementary Table S3 list the NCBI accession numbers of the 16S rRNA gene sequences.
Table 1.
The closest relatives, phylogenetic affiliations, phylotype clustering, and isolation origin of the 31 representative moonmilk isolates.
| Isolate | Closest relatives | 16S rRNA identity % (gaps) | Accession number | Origin of the closest relatives | COL | Md | Phylotype 16S → MLSA |
|---|---|---|---|---|---|---|---|
| MM1 | S. sp. CFMR 7 strain CFMR-7/S. fulvissimus DSM 40593 | 99.1 (4)/99.1 (6) | KU714864 | Plant (rubber)/unknown | COL3 | SN | I = I |
| MM3 | Un. bacterium clone Md-54/Un. bacterium clone 10–355 | 99.8 (0)/99.8 (0) | KU714892 | Soil/soil | COL3 | SN | II = II |
| MM5 | S. scabiei BCCO 10_524/S. europaeiscabiei 08-46-04-2 (#50) | 99.7 (0)/99.4 (2) | KU714904 | Both plant (potato) | COL3 | SN | III = III |
| MM6 | S. sp. Mg1/S. sp. SXY10 | 99.7 (0)/99.8 (0) | KU714910 | Glacier soil (Alaska)/soil | COL3 | SN | IV = IV |
| MM7 | S. sp. NEAU-spg16/S. sp. A42 | 99.6 (0)/99.9 (0) | KU714915 | Soil/soil | COL3 | SN | V = V |
| MM10 | S. sp. NEAU-QHHV11/S. sp. (Acc.Nr.D63866) | 99.1 (4)/98.6 (7) | KU714865 | Soil/soil | COL3 | SN | VI = VI |
| MM12 | S. sanglieri A14/S. sp. ME03-5656.2c | 99.8 (0)/99.6 (0) | KU714878 | Soil/plant (potato) | COL3 | SN | VII = VII |
| MM13 | S. turgidiscabies ATCC 700248/S. turgidiscabies WI04-05A | 98.8 (4)/98.7 (4) | KU714882 | Both plant (potato) | COL3 | SN | VIII = VIII |
| MM14 | S. anulatus strain 173826/S. anulatus strain 173541 | 100 (0)/100 (0) | KU714883 | Both unknown | COL3 | SN | IX = IX |
| MM17 | S. sp. Mg1/S. sp. SXY10 | 99.7 (0)/99.8 (0) | KU714885 | Glacier soil (Alaska)/soil | COL3 | SN | IV → XXVI |
| MM19 | S. sp. NEAU-spg16/S. sp. A42 | 99.6 (0)/99.9 (0) | KU714887 | Soil/soil | COL3 | SN | V → XXVII |
| MM21 | Un. bacterium clone Md-54/Un. bacterium clone 10–355 | 99.7 (0)/99.7 (0) | KU714888 | Soil/soil | COL3 | SN | XI = XI |
| MM23 | Un. bacterium clone Md-54/Un. bacterium clone 10–355 | 99.8 (0)/99.8 (0) | KU714890 | Soil/soil | COL3 | SN | II → XXVIII |
| MM24 | S. sp. ME02-6979.3a/S. sp. 1C-HV8 | 98.3 (5)/98.4 (4) | KU714891 | Plant (potato)/animals (ants) | COL3 | SN | XII = XII |
| MM44 | Un. bacterium clone Md-54/Un. bacterium clone 10–355 | 99.7 (0)/99.7 (0) | KU714900 | Soil/soil | COL3 | SN | XI → XXIX |
| MM48 | S. sp. HBUM171258/S. sp. Mg1 | 99.9 (1)/99.6 (0) | KU714903 | Unknown/glacier soil (Alaska) | COL3 | MMch | XIII = XIII |
| MM59 | S. sp. ID05-8D/S. sp. ID01-6.2a | 99.5 (1)/99.4 (1) | KU714909 | Both plant (potato) | COL3 | MMch | III → XXX |
| MM68 | S. turgidiscabies ATCC 700248/S. turgidiscabies WI04-05A | 99.0 (2)/99.0 (2) | KU714913 | Both plant (potato) | COL3 | B-4 | XIV = XIV |
| MM90 | S. sp. AA58/S. sp. AS40 | 99.5 (4)/99.4 (5) | KU714925 | Soil/soil | COL1 | ISP4 | XV → – |
| MM99 | S. fulvissimus DSM 40593/S. sp. ME02-6987.2c | 99.7 (2)/99.7 (2) | KU714928 | Unknown/plant (potato) | COL1 | ISP6 | XVI = XVI |
| MM100 | S. sanglieri A14/S. sp. ME03-5656.2c | 99.9 (0)/99.5 (0) | KU714866 | Soil/plant (potato) | COL1 | B-4 | XVII = XVII |
| MM104 | S. scopuliridis strain SCSIO ZJ46/S. sp. AK02-1a | 99.2 (0)/99.0 (0) | KU714869 | Deep sea/plant (potato) | COL3 | ISP6 | XVIII = XVIII |
| MM105 | S. finlayi strain CB00817/S. olivoviridis strain S3 | 99.4 (6)/99.3 (5) | KU714870 | Soil/animals (earthworm) | COL3 | ISP6 | XIX = XIX |
| MM106 | S. rishiriensis strain 1706/S. fimbriatus strain cfcc3155 | 99.0 (0)/98.8 (1) | KU714871 | Soil/unknown | COL3 | ISP1 | XX = XX |
| MM107 | S. pristinaespiralis strain HCCB 10218/S. sp. NEAU-bt10 | 98.8 (2)/98.8 (0) | KU714872 | Soil/soil | COL3 | ISP1 | XXI = XXI |
| MM108 | S. sp. SXY66/S. sp. 1H-TWYE2 | 100 (0)/99.3 (2) | KU714873 | Soil/animals (ants) | COL3 | ISP7 | XXII = XXII |
| MM109 | S. lunaelactis MM109T/S. lunaelactis MM15 | 100 (0)/99.9 (0) | KM207217.2 | Cave/cave | COL3 | ISP7 | X = X |
| MM111 | S. sp. 1H-TWYE2/S. sp. SXY66 | 99.7 (0)/99.5 (2) | KU714875 | Animals (ants)/soil | COL4 | ISP6 | XXIII = XXIII |
| MM117 | S. sp. PAMC26508/S. pratensis ATCC 33331 | 99.7 (0)/99.7 (0) | KU714876 | Antarctic lichen/soil | COL4 | ISP7 | XXIV = XXIV |
| MM122 | S. sp. PAMC26508/S. pratensis ATCC 33331 | 100 (0)/100 (0) | KU714879 | Antarctic lichen/soil | COL4 | B-4 | IX → XXXI |
| MM128 | S. sp. ZLN234/S. sp. SXY66 | 99.9 (0)/99.0 (4) | KU714881 | Glacier soil (Arctic)/soil | COL4 | SN | XXV = XXV |
B-4, B-4 agar medium; MMch, minimal medium with 1% chitin; ISP, International Streptomyces Project medium; SN, starch nitrate medium; COL, moonmilk collection site; Md, isolation medium; Un., uncultured. Symbols: -, isolates not included in the MLSA; T, Type strain (Maciejewska et al., 2015).
In order to profile the potential of moonmilk isolates to biosynthesize secondary metabolites, the genes coding for type I, type II, and type III polyketide synthases (PKS-I, PKS-II, and PKS-III) and non-ribosomal peptide synthetases (NRPS) were recovered from their genomes using antiSMASH v3.0.4 (Weber et al., 2015). Due to the fragmented nature of the genome assemblies (ranging from 318 contigs longer than 1 kb for MM23 to 1416 contigs for MM59) and to the large size of the modular NRPS and PKS-I genes (over 40 kb, Wang et al., 2014), the probability of finding complete coding sequences decreases together with the contig length. Therefore, counting the number of genes or clusters split across several shorter contigs would result in an overestimation of the total amount of such genes. To palliate the absence of fully assembled chromosomes while still collecting meaningful statistics, we decided to apply a cut-off on the length of the contigs selected for analysis (minimum length of 10 kb) and to consider NRPS and PKS-I gene sequences only when they displayed adenylation and acyltransferase domains, respectively. These domains, which are the highly selective gatekeeper enzymes for the incoming monomeric building blocks, are required for the initiation and elongation modules of the NRPS/PKS-I clusters. The number of predicted genes of each category for each individual phylotype is compared to their respective mean antimicrobial activities against Gram-positive, Gram-negative bacteria, and fungi. The accession numbers of NRPS and PKS-I/II/III genes are listed in Supplementary Table S4.
Phylogenetic Analysis
To carry out phylogenetic analysis, in each nucleotide alignment of the six selected housekeeping genes (see above), the sequences from the three reference strains used to mine the moonmilk genomes were removed. Then, positions with missing character states in >5 moonmilk isolates were removed. Finally, the trimmed alignments were concatenated into a single (MLSA) supermatrix of 10,632 nucleotides for 70 isolates using SCaFoS v1.30k (Roure et al., 2007). For 16S rRNA phylogenies, the closest relatives to the moonmilk isolates were recovered by BLAST searches using full-length 16S rRNA sequences, along with the Streptomyces isolates from a moonmilk deposit in Siberia (Axenov-Gribanov et al., 2016). For the eight isolates for which the genomes were not sequenced (MM9, MM32, MM39, MM55, MM73, MM88, MM90, MM93), nearly full-length 16S rRNA sequences were obtained using PCR primers and conditions as previously reported (Maciejewska et al., 2015). The 78-moonmilk strain alignment of the 16S rRNA was further processed using the software “two-scalp” (D. Baurain, to be published elsewhere) to integrate 35 additional sequences, corresponding to the 27 (non-redundant) best BLAST hits, 7 Siberian moonmilk sequences, and the sequence of Saccharopolyspora erythraea, used as the outgroup.
Both the MLSA supermatrix and the 16S rRNA alignment were submitted to phylogenetic inference using the rapid bootstrap analysis of RAxML v8.1.17 (Stamatakis, 2014; 100 pseudoreplicates) under the model GTR+I+Γ4. The resulting MLSA and 16S rRNA trees were first formatted in FigTree v1.4.21 then further arranged using Inkscape v0.912. Patristic distances between moonmilk isolates were derived from the MLSA tree using the TREEPLOT program of the MUST software package (Philippe, 1993).
Antimicrobial Activity Screening
Antimicrobial activities of one representative of each phylotype deduced from the MLSA, together with MM90 (representing 16S-phylotype XV) were tested using the cross-streak method on two (antifungal test) to five (antibacterial test) different culture media: Mueller Hinton Agar (MHA) (Difco, BD, Franklin Lakes, NJ, USA), Tryptic Soy Agar (TSA) (tryptone 15 g, soybean meal 5 g, NaCl 5 g, agar 15 g; pH 7.3), ISP media No. 7 (Shirling and Gottlieb, 1966), starch nitrate (SN) medium (Waksman and Lechevalier, 1961), and minimal medium (Kieser et al., 2000) supplemented with 25 mM N-acetylglucosamine (MM + GlcNAc). Each moonmilk strain was independently inoculated from the mycelium stock as a single line in the center of the square plate and incubated for 7 days at 28°C, before being cross-streaked with bacterial or fungal reference strains.
Antibacterial activities were tested against a range of Gram-positive and Gram-negative bacteria, including Escherichia coli (ATCC 25922), Pseudomonas aeruginosa (ATCC 27853), Citrobacter freundii (ATCC 43864), Klebsiella pneumoniae (ATCC 13883), Bacillus subtilis (ATCC 19659), Staphylococcus aureus (ATCC 25923), and Micrococcus luteus (ATCC 9341). Each tested bacteria was cross-streaked perpendicular to the growth of the moonmilk isolate at the distance of 2 cm from one another from a suspension prepared according to EUCAST recommendations (EUCAST, 2015). Briefly, each inoculum was made from the overnight-grown plate of each pathogen in solid LB (Difco, BD, Franklin Lakes, NJ, USA) at 37°C by suspending several morphologically similar colonies in LB broth (Difco, BD, Franklin Lakes, NJ, USA) until the OD625 nm reached 0.08–0.13, corresponding to the McFarland 0.5 standard. The reference bacterial strains were cross-streaked with a cotton swab, the plates incubated overnight at 37°C, and the activities determined by measurement of the inhibition zone (in cm).
Antifungal activities were tested against a range of pathogenic fungi including Candida albicans (ATCC 10231), C. albicans (azole-resistant routine strain from the National Reference Center for Mycoses, 13-160409-5014), referred as C. albicans ‘R,’ Aspergillus fumigatus (Neqas 1210), Rasamsonia argillacea (Neqas 1872), Penicillium chrysogenum (Neqas 2068), and Trichophyton mentagrophytes (Neqas 1208). Each fungal strain was suspended in water at the density equivalent to 0.5 McFarland, and the obtained fungal suspension was perpendicularly cross-streaked against moonmilk isolates with a distance of 4 cm from one another. The plates were incubated at 37°C, for up to 4 days, and the measurements of inhibition zones (in cm) were recorded every day.
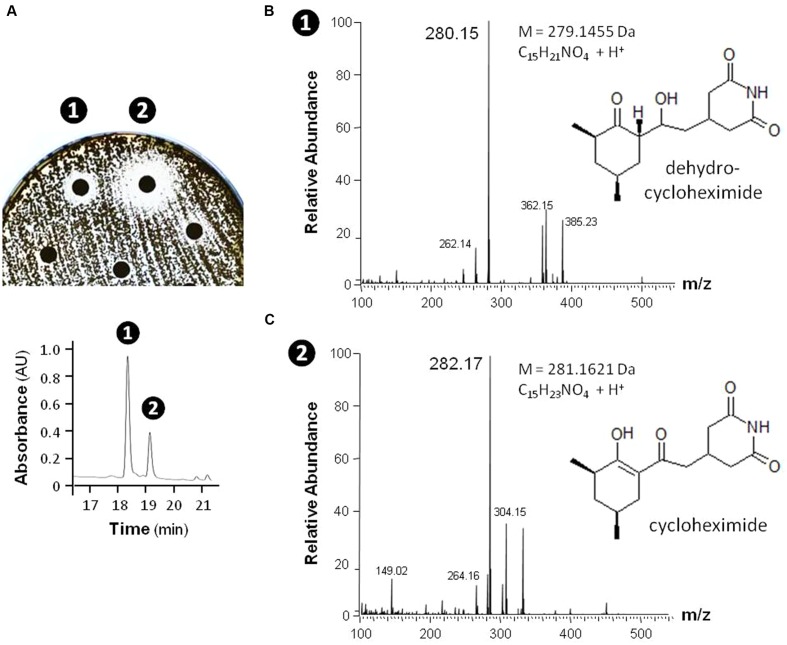
MM99 Isolate Antifungal Agents Extraction and Purification by High Pressure Liquid Chromatography (HPLC)
Streptomyces sp. MM99 was inoculated on 15 Glucose Yeast and Malt medium (GYM; glucose 4 g; yeast extract 4 g; malt extract 10 g; casein enzymatic hydrolysate 1 g; NaCl 2 g; agar 20 g) plates and incubated for 10 days at 28°C. The agar was collected, poured into a flask with an equal volume of ethyl acetate (∼300 ml) and agitated overnight at room temperature for metabolites extraction. Ethyl acetate was collected and pieces of agar were removed by centrifugation (25 min at 4000 rpm) before the solvent was evaporated on a rotary evaporator (IKA RV10 digital, VWR, Radnor, PA, USA). The dried crude extract was resuspended in 4 ml of pure methanol high pressure liquid chromatography (HPLC Barker UHPLC grade). Prior to fractionation by HPLC, the antifungal activity of the total extract was assessed by a disk diffusion assay on a yeast peptone dextrose (YPD; peptone 20 g; yeast extract 10 g; glucose 20 g; agar 15 g) agar plate inoculated with Saccharomyces cerevisiae (ATCC 9763; with a cotton swab dipped in a 0.25–0.27 OD625 LB suspension). The full extract was then fractionated by HPLC (Waters, Milford, MA, USA) using a Waters 2695 Separations Module (Alliance) with a Waters 2998 Photodiode Array Detector coupled to a Waters Fraction Collector WFC III. The methanol extract was analyzed on a Nucleodur C18ec column (2.0 mm × 150 mm, 5 μm particle size, Macherey-Nagel) at a column temperature of 40°C. Extract separation was achieved by increasing the acetonitrile (Barker, HPLC far UV grade)/water (milliQ filtrated on 0.22 μm) + 0.05% trifluoroacetic acid (TFA, Sequencing grade; Thermo Fisher Scientific, San Jose, CA, USA), ratio (from 0 to 62.5% of acetonitrile during 30 min, then from 62.5 to 100% of acetonitrile during 8 min) at a 300 μl/min flow rate. Online UV absorption measurement was performed from 190 to 800 nm. Data were analyzed using Empower 3 software (Waters, Milford, MA, USA). The obtained extract fractions were subsequently tested for antifungal activities by a disk diffusion assay as described above.
Compound Identification by Ultra-Performance Liquid Chromatography-Tandem Mass Spectrometry (UPLC-MS/MS)
Fractions displaying antifungal activities revealed by the disk diffusion assay were analyzed by liquid chromatography–tandem mass spectrometry (LC–MS/MS). Briefly, compounds were separated by reverse-phase chromatography using Ultra Performance Liquid chromatography (UPLC IClass, Waters) using a Nucleodur C18ec column (2.0 mm × 150 mm, 5 μm particle size, Macherey-Nagel). Elution was achieved by increasing the acetonitrile/water (milliQ filtrated on 0.22 μm) + 0.05% trifluoroacetic acid ratio (from 0 to 62.5% during 30 min, then from 62.5 to 100% during 8 min) at a 300 μl/min flow rate. On-line UV absorption measurement was performed at 210 and 265 nm and the chromatography system was finally coupled to a Q Exactive Plus hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Fisher Scientific, San Jose, CA, USA), operated in positive ion mode and programmed for data-dependent acquisitions. Survey scans were acquired at mass resolving power of 140,000 FWHM (full width at half maximum) from 100 to 1500 m/z (1 × 106 ions accumulation target). The five most intense ions were then selected to perform MS/MS experiments by Higher Energy Collision Dissociation (HCD) fragmentations using stepped normalized collision energy (NCE; 21,2; 25; 28) within 2 amu isolation windows (resolution 17500, 1 × 105 ions accumulation target). A dynamic exclusion was enabled for 10 s. Data were analyzed using Xcalibur v2.2 (Thermo Fisher Scientific, San Jose, CA, USA). Commercial cycloheximide standard (Sigma-Aldrich, St. Louis, MO, USA) was used as a control.
Results and Discussion
Isolation of Actinobacteria from Moonmilk Deposits
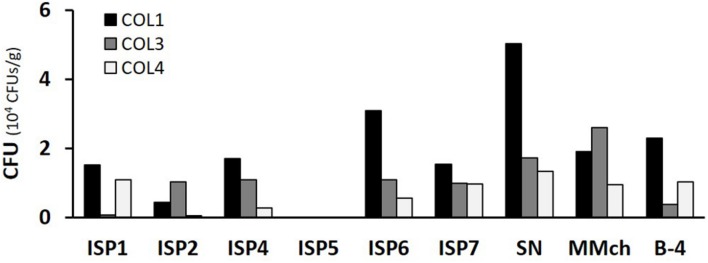
Cultivable actinobacterial species were isolated from the three separate moonmilk deposits within Grotte des Collemboles (Figure 1). Our cultivation-based screening on Actinobacteria-specific media revealed high population density across the three collection sites studied, with up to 104 CFUs/g (Figure 2). No microbial growth could be detected in the ISP5 medium (Glycerol Asparagine Agar), while the highest numbers of CFUs for each sampling point were recorded on the starch nitrate (SN) medium (5 × 104 and 1.3 × 104 CFUs/g in COL1 and COL4, respectively), and on the minimal medium supplemented with chitin (2.6 × 104 CFUs/g in COL3; Figure 2). Most of the viable isolates (>50%) were chosen from the starch nitrate medium (Table 1; Supplementary Table S3), in which the most distinct colony phenotypes were observed. From the 129 colonies initially selected, only 78 were recovered as pure isolates after three rounds of inoculation/cultivation, and were preserved for further studies. This important loss (40%) during the purification steps was expected, as living in an oligotrophic environment necessarily implies nutrient exchange between individuals of the microbiome, a cooperative strategy that was only possible in the non-diluted plates where colonies were originally picked. Inoculation of single colonies thus biased the selection of specimens able to feed only from nutrients present in the synthetic medium, whereas species most adapted to cooperative growth certainly represented a significant part of the isolates lost during the screening procedure.
FIGURE 2.
Total number of CFU obtained from each moonmilk collection point according to the selective media used. CFU, colony-forming units; ISP, International Streptomyces Project; SN, starch nitrate; MMch, minimal medium supplemented with chitin.
Phylogeny of Cultivable Moonmilk-Derived Streptomyces Isolates
When cultivated in liquid cultures, all the isolates formed filamentous pellets, suggesting their affiliation to the genus Streptomyces or to other filamentous Actinobacteria (data not shown). BLAST search using either full or nearly full-length 16S rRNA gene sequences indeed revealed that the closest hits to each of the moonmilk-derived isolate were Streptomyces species displaying between 98.3 and 100% identity (Table 1; Supplementary Table S3). Cultivable members of this genus have been previously reported from moonmilk (Cañaveras et al., 1999; Axenov-Gribanov et al., 2016), however isolates belonging to other genera, such as Amycolatopsis, Saccharothrix, Acinetobacter, Chryseomonas (Cañaveras et al., 1999), Arthrobacter (Rooney et al., 2010), Pseudonocardia, Propionibacterium (Portillo and Gonzalez, 2011), and Nocardia (Axenov-Gribanov et al., 2016), have been found through both culture-dependent and independent approaches. It was therefore unexpected that only Streptomyces were isolated across a wide range of Actinobacteria-specific media. One reason for such biased recovery might be that Streptomyces are saprophytes accustomed to use a large variety of nutrient sources, and are thus better adapted to grow on the synthetic rich media used in our screening procedure. Secondly, among actinobacterial populations, Streptomyces are known for their more rapid growth in comparison to other genera, recognized as rare Actinobacteria, which are isolated much less frequently (Subramani and Aalbersberg, 2013). Therefore, in our in vitro culture conditions, Streptomyces have probably overtaken nutrients and space over other, slower growing, actinobacterial representatives.
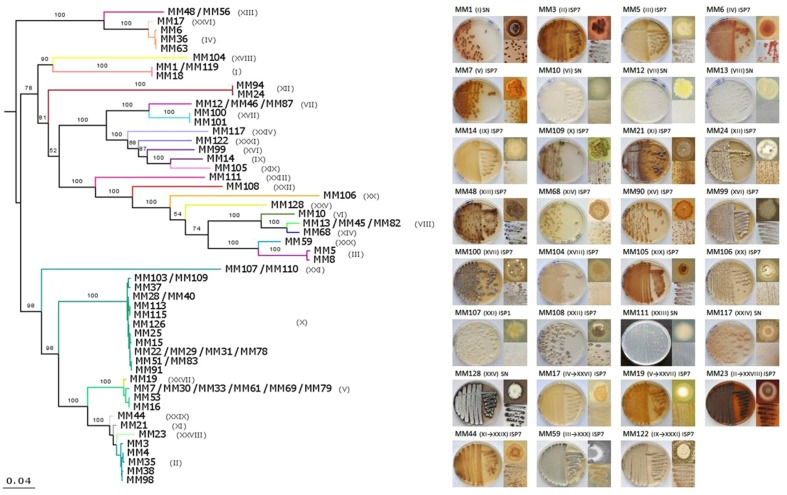
Based on the 16S rRNA tree topology, we grouped the 78 moonmilk isolates into 25 phylogenetic clusters (Figure 3; Table 1; Supplementary Table S3). The largest cluster on the tree (Figure 3, phylotype X), delineating the recently described Streptomyces lunaelactis species, grouped six novel isolates in addition to the 13 strains reported previously (Maciejewska et al., 2015), which collectively originate from each of the studied sampling site. The clustering of isolates from multiple sampling sites in a single branch was also observed for six other phylotypes, namely phylotypes I, II, VII, IX, XI, and XII, which suggests a widespread occurrence of these isolates within the moonmilk deposits of the cave.
FIGURE 3.
16S rRNA-based phylogenetic tree of moonmilk cultivable isolates (MM). The collection points, from which each moonmilk isolate originates are indicated by colored symbols, while the phylotype affiliations are indicated in brackets. The Streptomyces isolates originating from moonmilk deposits of the Bolshaya Oreshnaya Cave in Siberia are marked by a star. The alignment of nearly complete 16S rRNA sequences had 1413 unambiguously aligned nucleotide positions for 113 strains. The evolutionary model was GTR+I+Γ4 and bootstrap values are based on 100 pseudoreplicates (the bootstrap values below 40% are not displayed). Saccharopolyspora erythraea was used as outgroup, and its branch was reduced five times which is indicated by the slanted bars. The scale bar represents 0.05 substitutions per site.
Among the closest environmental Streptomyces species deduced from the 16S rRNA BLAST searches, 36 (46%) have been isolated from soils, 12 (15%) from plants, while relatives associated with water ecosystems, lichens, and animals constituted 2 (3%), 2 (3%), and 1 (1%), respectively (Table 1; Supplementary Table S3). Finally, 19 isolates (24%) were found to be affiliated to the recently described moonmilk species, S. lunaelactis (99.9 or 100% 16S rRNA identity), with isolate MM109T selected as the type strain (Maciejewska et al., 2015). The closest known relative of S. lunaelactis found through full-length 16S rRNA gene search, Streptomyces globisporus, further increases the number of relatives originating from soils to 55 (71%). The high sequence identity of 16S rRNA genes of moonmilk isolates to species from soil, plant, and water environments suggests that the moonmilk may have been seeded with Actinobacterial species brought into the cave from the surface by dripping water (Laiz et al., 1999; Legatzki et al., 2012; Yun et al., 2015). It should be noted that some of our phylotypes are phylogenetically closely related to Streptomyces isolates recently identified from moonmilk deposits of the Bolshaya Oreshnaya Cave in Siberia (Axenov-Gribanov et al., 2016). This is particularly the case for phylotypes IX, X, XVIII, and XXIV (Figure 3), however the comparison is restricted by the small size of the collection generated from the Siberian cave (limited to seven Streptomyces isolates). Nonetheless, this observation supports the idea that we may have identified at least some bacterial species specifically associated with moonmilk speleothems.
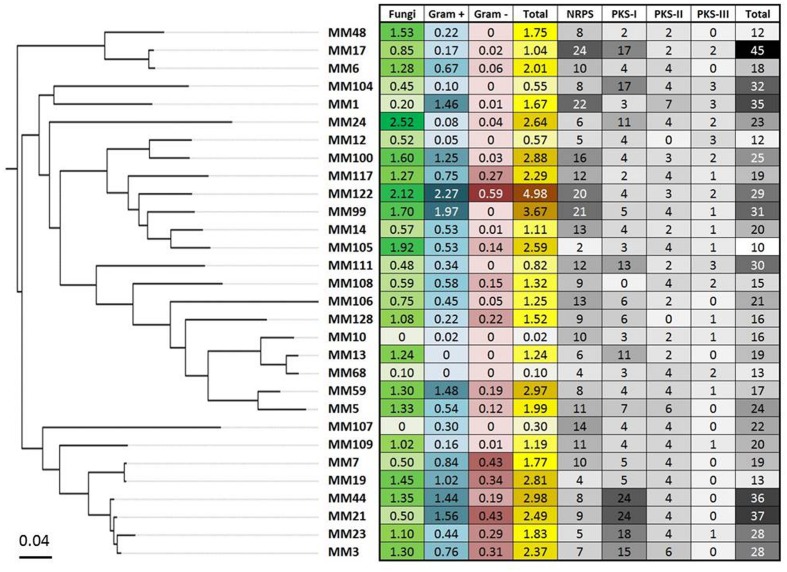
As 16S rRNA-based phylogeny is known to be insufficient for discriminating between closely related Streptomyces species (Guo et al., 2008; Labeda, 2011; Rong and Huang, 2012) and, consequently, could underestimate the true diversity of our collection, a MLSA was additionally performed. Instead of partial gene fragments used previously (Maciejewska et al., 2015), we have retrieved nearly full-length sequences of the selected housekeeping genes (atpD, rpoB, trpB, gyrB, and recA) from the genomes of 70 out of 78 isolates. As expected, the phylogenetic resolution of the MLSA tree, based on a supermatrix of 10,632 unambiguously aligned nucleotides (including 16S rRNA), was much higher. Hence, some strains previously considered to belong to a single phylotype were separated into several sub-clusters (Figure 4A). To clearly delineate the phylotypes, the comparison of the pairwise patristic distances between isolates was taken into account together with the tree topology (Supplementary Figure S1; Figure 4A). The highest pairwise distance among S. lunaelactis strains (0.0096) was considered as the threshold for phylotype assignment since most of these isolates were previously reported to belong to a single species (Maciejewska et al., 2015). Consequently, the number of phylogenetic clusters increased from 25, inferred from the 16S rRNA tree, to 31 based on MLSA. The new phylotypes differentiated through MLSA were represented by MM17 (IV→XXVI), MM19 (V→XXVII), MM23 (II→XXVIII), MM44 (XI→XXIX), MM59 (III→XXX), and MM122 (IX→XXXI) (Table 1). To assess whether the distinction of new phylotypes was supported by phenotypic traits, we compared pigment production and colony morphologies between representative isolates of MLSA phylotypes and representative isolates of their former 16S rRNA-based phylotypes. For instance, the separation of isolate MM17 (now phylotype XXVI) from isolates of phylotype IV (represented by MM6) was indeed additionally supported by the phenotype analysis, as unlike MM6 isolate, which produces a dense pink-reddish intracellular pigment, isolate MM17 remains unpigmented when grown on ISP7 (Figure 4B). Dissimilar phenotypes were also observed between representative isolates that formerly belonged to the same phylotype, i.e., MM7 and MM19, MM23 and MM3, MM44 and MM21, MM59 and MM5, and MM122 and MM14 (Figure 4B).
FIGURE 4.
Multilocus sequence analysis (MLSA) phylogeny of moonmilk isolates and phenotypes of the selected representative of each phylotype. (A) MLSA was based on the full-length sequences of six housekeeping genes (16S rRNA, atpD, recA, rpoB, gyrB, trpB). Each MLSA-deduced phylotype is indicated by a specific color and the corresponding number in brackets. The supermatrix had 10,632 unambiguously aligned nucleotide positions for 70 isolates. ML phylogenetic inference was carried out as for 16S rRNA alone. The scale bar represents 0.04 substitutions per site. (B) Phenotype of the 31 phylotype representative isolates. The MLSA phylotype number is indicated in brackets, with the former, 16S rRNA-deduced phylotypes included. For each isolate, front and back of the Petri dish-grown bacteria are presented together with the phenotype of a single colony.
Antimicrobial Activity Screening
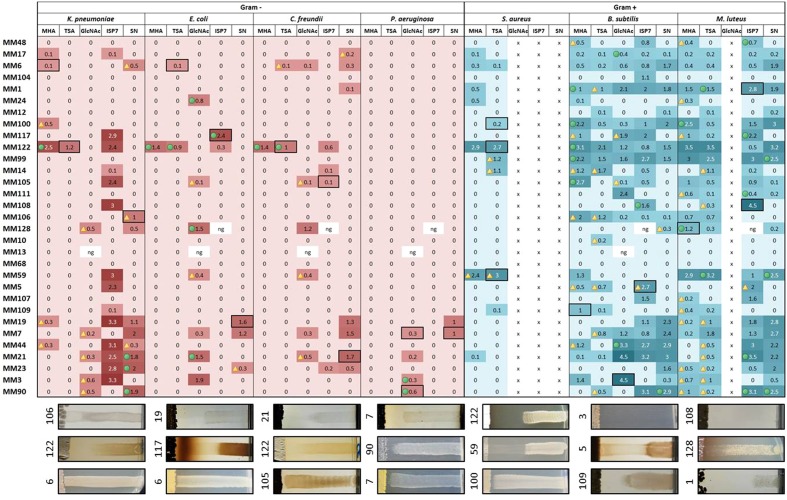
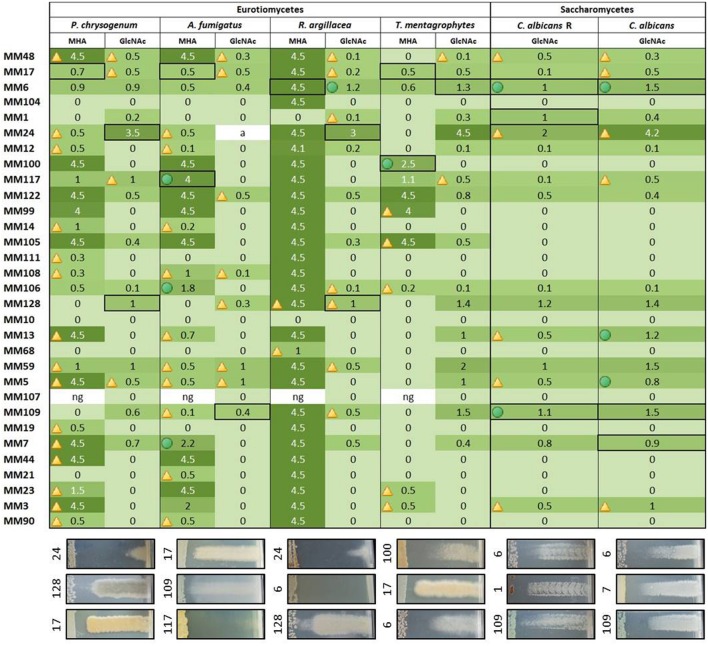
The potential for antimicrobial activity of the moonmilk-derived cultivable isolates was assessed for one representative of each of the 31 phylotypes deduced from the MLSA (see heatmaps in Figures 5 and 6 for antibacterial and antifungal activities, respectively). Two categories of antimicrobial activities were recorded, i.e., (i) those that fully inhibited the growth (GI, growth inhibition) of the tested reference strains (see Figure 5 MM122 against S. aureus as an example), and (ii) those that allowed only partial growth (IG, impaired growth, see Figure 5 MM5 against B. subtilis as an example).
FIGURE 5.
Heatmap illustrating the strength of the antibacterial activities of the representatives of moonmilk phylotypes. The size of the inhibition zone (in cm) is indicated for each moonmilk isolate and related to a color scale. Isolates causing impaired growth of the tested bacteria are indicated by a yellow triangle, inhibition effects combining both impaired growth and growth inhibition are marked with a green circle, and clear bactericidal growth inhibition effects are not marked with any symbol. The fields selected with a black border refer to the chosen examples of the cross-streak results displayed below the heatmap. x, the tested pathogen cannot grow in the chosen medium; ng, no or poor growth of the moonmilk isolate.
FIGURE 6.
Heatmap illustrating the strength of the antifungal activities of the representatives of moonmilk phylotypes. The size of the inhibition zone (in cm) is indicated for each moonmilk isolate and related to a color scale. Isolates causing impaired growth of the tested fungal strains are indicated by a triangle, inhibition effects combining both impaired growth and growth inhibition are marked with a green circle, and clear bactericidal growth inhibition effects are not marked with any symbol. The fields selected with a black border refer to the chosen examples of the cross-streak results displayed below the heatmap. ng, no or poor growth of the moonmilk isolate.
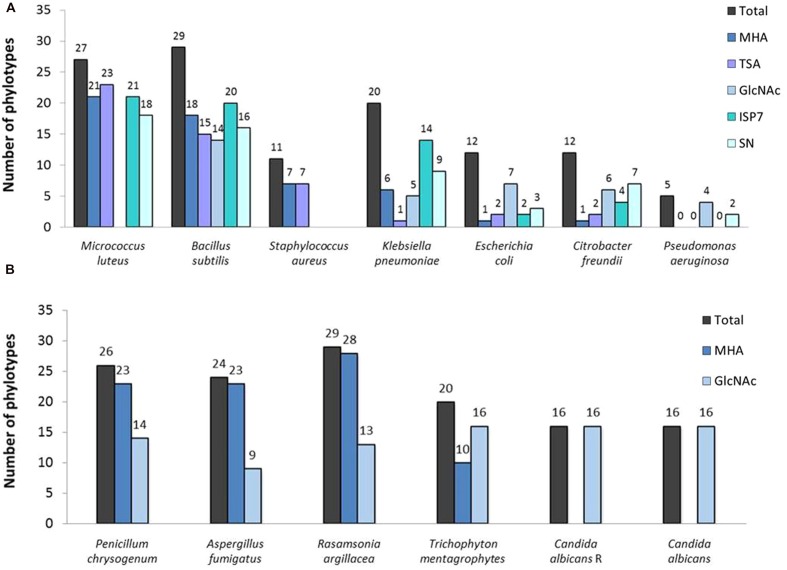
The screening for antibacterial properties was carried out under five different culture conditions, i.e., the two complex and nutrient rich media (MHA and TSA), the ISP7 and SN media from which most of our strains were isolated (see Table 1; Supplementary Table S3), and the minimal medium supplemented with GlcNAc, which is a known elicitor of antibiotics under poor culture conditions (Rigali et al., 2008; Światek et al., 2012; Zhu et al., 2014). Results from the antibacterial cross-streak are presented in the heatmap of Figure 5 (with particular cases highlighted in Figure 7), and summarized in Figures 8A and 9. Overall, the moonmilk isolates expectedly displayed a much stronger antibacterial activity (GI and IG) against Gram-positive bacteria (94% of the phylotypes) than against Gram-negative bacteria (71% of the phylotypes). Indeed, 94% of the tested isolates were active against B. subtilis, 87% against M. luteus, and 36% against S. aureus, while 65% were found to show activity against K. pneumoniae, 39% against E. coli, 39% against C. freundii and only 16% against P. aeruginosa (Figures 5 and 8A). The most active isolate in terms of both strength and spectrum of targeted pathogens was MM122 (Figures 5 and 9). Interestingly, a group of four isolates (MM3, MM7, MM21, and MM90) similarly triggered the production of compounds active against Gram-negative bacteria, particularly P. aeruginosa and K. pneumonia, in the presence of GlcNAc (Figure 5). All these isolates branch together in the 16S rRNA and the MLSA-deduced phylogenetic trees (Figures 3 and 4A), which suggests that their similar response to the GlcNAc eliciting molecule should be induced by the same signaling pathway and involves similar or identical bioactive compounds. Those isolates together with other closely related moonmilk strains, namely MM19, MM44, and MM23, showed one of the strongest inhibitory profiles against a broad spectrum of pathogens and under most of the tested culture conditions. This suggests that they possibly might share a similar set of antibacterial secondary metabolite clusters. In contrast, MM10, MM13, and MM68, all representatives of closely related phylotypes, did not reveal any antibacterial activity; however, genome mining showed that those three isolates are predicted to possess in total 16, 19, and 13 biosynthetic cluster-associated genes (including NRPS, all types of PKS), respectively (Figure 9). This suggests that the absence of antibiotic activity would be rather caused by inappropriate culture conditions for eliciting the production of cryptic antibiotics, rather than a lack of genetic material associated with secondary metabolism.
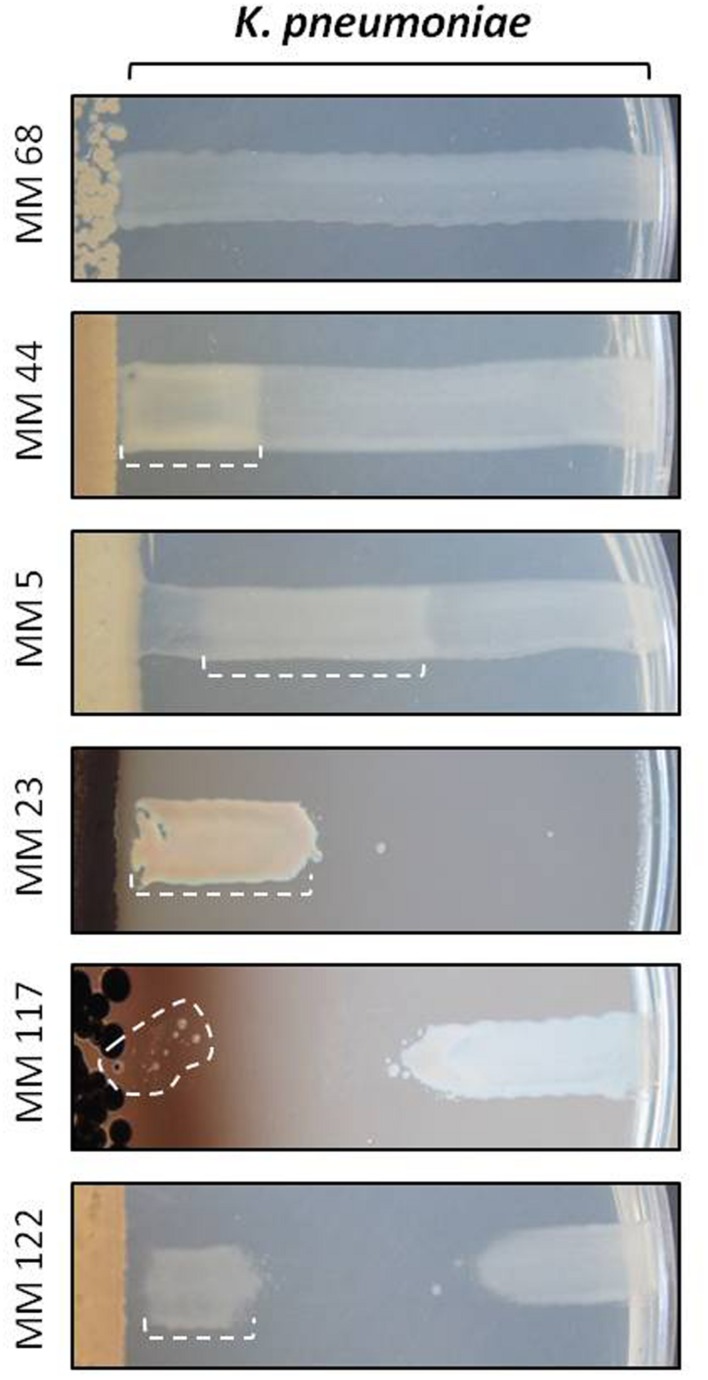
FIGURE 7.
Illustration of the paradoxical zone phenomenon observed with Klebsiella pneumoniae. Zones with the so-called Eagle or hormesis effects are delineated by a dotted line. Note that for compounds produced by isolate MM117 we only observed the emergence of isolated resistant colonies instead of the wide and confluent zone of growth observed for the other marked cases. MM68 was chosen as a control to show non-affected growth of K. pneumoniae on the ISP7 medium.
FIGURE 8.
Total number of the representatives of the phylotypes displaying an activity against (A) bacterial and (B) fungal strains under each tested condition.
FIGURE 9.
The global antimicrobial pattern of moonmilk Streptomyces isolates and their presumed secondary metabolites biosynthesis potential based on genome mining. The heatmap plot, correlated with MLSA-based phylogeny, represents the mean of the antifungal and antibacterial (Gram-positive and Gram-negative bacteria) activities for each of the 30 phylotype representatives, together with the sum of those activities. In addition, the greyscale on the heatmap displays the biosynthetic genes content of individual strain, including number of its NRPS, PKS-I, PKS-II, and PKS-III genes, together with their total number.
Another remarkable example evidencing that the antibiotic production highly depends on the culture medium composition is illustrated by the isolate MM108, which did not display any antibacterial activity unless grown on the ISP7 medium, where it induced a strong GI effect against K. pneumonia, B. subtilis, and M. luteus (Figure 5).
Strikingly, the ISP7 medium also stimulated unusual growth phenotypes by K. pneumoniae to compounds produced by our moonmilk isolates. All different atypical cases observed for this pathogen are presented in Figure 7. For instance, beyond the classical GI and the IG effects (see Figure 5), we observed that K. pneumoniae would grow or be only slightly inhibited near the antimicrobial producing strain, whereas its growth was fully or partially inhibited (for example MM23 in Figure 7) or impaired (for example MM5 in Figure 7) at a higher distance. This non-linear response to a diffusion gradient of the secreted antibiotics has been described as the Eagle effect (Eagle and Musselman, 1948). According to this phenomenon, the activity of antimicrobial compounds is paradoxically higher at lower concentrations, whereas high concentrations allow growth within the vicinity of Streptomyces. The observed growth close to the antimicrobial-producing strain might be due to the onset of the resistance mechanisms, which were reported to be rapidly induced by some antibiotics in K. pneumoniae (Zhong et al., 2013; Adams-Sapper et al., 2015). Indeed, several resistant K. pneumoniae colonies were detected in the proximity to the moonmilk isolate MM117 (Figure 7). Another possible explanation might be that the high production of antimicrobials by certain moonmilk isolates on the ISP7 medium could lead to precipitation and consequently inactivation of the antibacterial agents active against K. pneumoniae. Finally, the observed atypical response might be explained by a certain form of the hormesis phenomenon, which states that ‘the dose makes the poison’ (Zhanel et al., 1992; Linares et al., 2006; Allen et al., 2010). Consequently, the produced molecules would effectively suppress the growth of the pathogen, but only at the appropriate concentration ranges (Migliore et al., 2013).
Our collection of cultivable moonmilk Streptomyces strains was further assessed for the production of compounds with antifungal activities (Figure 6). The corresponding screening was carried out under two different culture conditions, i.e., (i) the rich medium MHA and (ii) the minimal medium supplemented with GlcNAc. The latter medium was again selected as GlcNAc, being the monosaccharide composing chitin, exerts a strong carbon catabolite repression on the induction of the chitinolytic system in Streptomyces species (Saito et al., 2007; Colson et al., 2008). This prevents ‘false-positive’ growth inhibition of tested fungi due to enzymatic hydrolysis of their chitin-based cell wall. The results of antifungal screening are presented in Figure 6 and summarized in Figures 8B and 9. Overall, the antifungal activities were stronger when the Streptomyces strains were grown on the rich MHA medium (Figures 6 and 8B). Globally, 94% of the tested isolates showed inhibitory activities against fungal strains, with 84% being specifically active against P. chrysogenum, 77% against A. fumigatus, 94% against R. argillacea, 65% against T. mentagrophytes, and 52% active against both C. albicans and its drug-resistant strain, C. albicans ‘R’ (Figures 6 and 8B). The most bioactive isolate in terms of the strength and the number of inhibited fungal strains was found to be MM24, while MM10 and MM107 did not show any suppression under conditions tested (Figure 6). The latter two isolates, together with MM68, generally represented the least prolific antimicrobial producers (Figure 9). The correlation between phylogenetic inference and detected activities was significant for moonmilk phylotypes MM6 and MM17, as well as MM5 and MM59 (Figure 6). Remarkably, most of our isolates displayed very strong growth inhibitory properties against the clinically relevant filamentous fungi – R. argillacea. This mold, belonging to the so-called R. argillacea species complex, is a causative agent of chronic infections of cystic fibrosis (CF) and chronic granulomatous disease (CGD) patients, displaying tolerance to various antifungals (Giraud et al., 2013). That almost all our isolates display high GI activity against R. argillacea suggests the involvement of one or more molecules commonly produced by cultivable moonmilk streptomycetes. The identification of active compound(s) is currently under investigation.
In order to provide a first evidence that the observed antimicrobial activities are indeed caused by the production of small inhibitory molecules, and do not result from other factors, such as enzymatic degradation, a single isolate – MM99 – was selected to characterize the compound(s) responsible for its antifungal response. For this purpose, MM99 was cultivated on GYM media, which triggered its antifungal activity, with the media collected for metabolite extraction and separation by HPLC. The two most active HPLC fractions were collected (Figure 10A), and subjected to UPLC-ESI-MS/MS analysis, which identified two masses (281.1626 and 279.1470 Da). These two peaks corresponded to cycloheximide and its precursor, dehydrocycloheximide, respectively (Figures 10B,C). Cycloheximide is a known inhibitor of eukaryotic protein synthesis, which was first isolated from Streptomyces griseus and previously known as actidione (Whiffen et al., 1946; Leach et al., 1947). To additionally confirm that our newly isolated antifungal compound was indeed cycloheximide, we compared the MS/MS fragments profile between the compound isolated from MM99 (Figure 10C) and a cycloheximide standard. Identical fragmentation patterns of both samples (Supplementary Figure S2), ultimately confirmed that cycloheximide was responsible – at least in part – for the antifungal activity of the isolate MM99. The identification of this single molecule is representative of ongoing efforts that aim to exhaustively identify the bioactive molecules of cultivable Streptomyces from moonmilk deposits.
FIGURE 10.
Identification of cycloheximide and dehydrocylcoheximide as antifungal compounds produced by the isolate MM99. The antifungal activities of MM99 were detected through disk diffusion assay (A, top) of HPLC-separated (A, bottom) fractions of the crude metabolites extract. The two bioactive fractions detected were subsequently identified by UPLC-MS/MS to contain mass peaks corresponding to dehydrocycloheximide (B) and cycloheximide (C).
Genome Mining
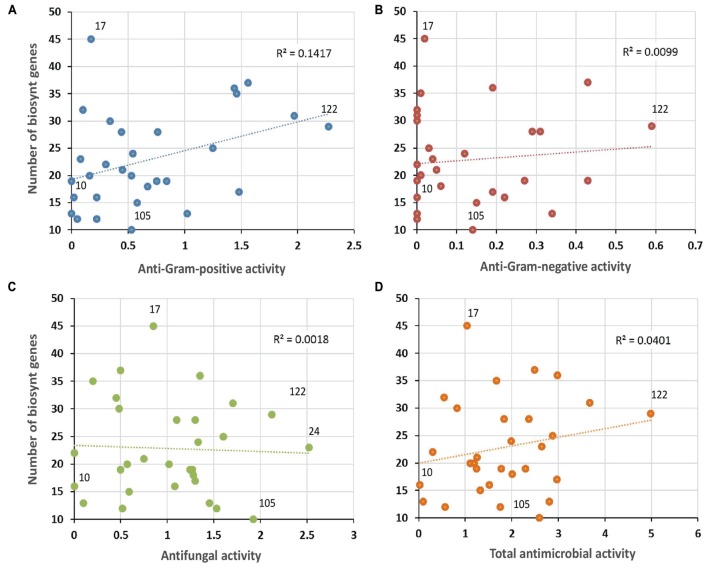
Complementary to in vitro antimicrobial screenings, we have carried out in parallel a large scale genome mining investigation. For this purpose, the draft genomic assemblies were examined for the presence of biosynthetic genes encoding polyketide synthases – PKSs (including PKS-I, PKS-II, and PKS-III), as well as NRPSs, which are collectively responsible for production of bioactive molecules in Actinobacteria. Notably, 100% of evaluated strains encoded at least two NRPS genes, 97% were found to possess PKS-I, 94% PKS-II, and 48% PKS-III genes (Figure 9). This clearly indicates a significant predisposition of moonmilk isolates to secrete bioactive secondary metabolites. The isolate MM17 was found to carry the highest total number of biosynthetic genes (45), while MM105 harbored only 10 of them, being classified as the weakest potential antimicrobial producer (Figure 9). Comparison of the biosynthetic genes content and the antimicrobial activities of each individual phylotype did not show significant correlation, with only small relationship observed for anti-Gram positive test (Figure 11). Remarkably, MM17 although encoding the highest amount of secondary metabolites genes displayed one of the weakest antimicrobial activities (Figure 11). This observation clearly highlights the urgency for finding appropriate culture conditions and triggers which would awaken silent metabolite clusters.
FIGURE 11.
Correlation between antimicrobial activities and predicted secondary metabolite biosynthetic genes. The vertical axis refers to the sum of PKS-I, PKS-II, PKS-III, and NRPS biosynthetic genes identified in each phylotype representative by the antiSMASH software. Horizontal axes represent the mean of the antibacterial activity (against Gram-positive bacteria (A), or Gram-negative bacteria (B), the antifungal activity (C), and the total antimicrobial activity (D) of each phylotype representative. Note the non-significant R-squared (R2) values for each plot, which reflect weak correlation between antimicrobial activities and the genetic potential of each isolate to produce secondary metabolites. The isolates that displayed the lowest (MM10) and the highest antibacterial (MM122) and antifungal (MM24) activities, and the smallest (MM105) and the largest (MM17) numbers of secondary metabolite biosynthetic genes, are highlighted in the plots.
Conclusion
In this work we have isolated 78 cultivable Streptomyces strains from three moonmilk deposits collected in the Grotte des Collemboles, Comblain-au-Pont (Belgium), and assessed their capacity to secrete compounds with antibacterial and antifungal activities. Surprisingly, the isolated strains were found to exclusively belong to the single Streptomyces genus, despite other cultivable-dependent and -independent approaches revealed the presence of many other Actinobacteria in moonmilk speleothems (Cañaveras et al., 1999; Rooney et al., 2010; Portillo and Gonzalez, 2011; Axenov-Gribanov et al., 2016). Hence, a more specific protocol for the isolation of rare Actinobacteria is currently tested in order to assess their potential in producing bioactive natural compounds. Phylogenetic analysis revealed the novelty of the selected isolates, and suggests that they might indeed represent indigenous moonmilk populations adapted to life in an inorganic and oligotrophic environment. Overall, antimicrobial screening showed that moonmilk strains were much more active against fungi, than against the bacterial reference strains, which was previously observe for plant-associated actinobacterial isolates (Bascom-Slack et al., 2009; Verma et al., 2009; Zhao et al., 2011). Identification of bioactive compounds is under investigation, particularly molecules active against R. argillacea. A complementary, in silico genome mining approach additionally revealed a high richness and diversity of secondary metabolite gene clusters, as evidenced by the presence of numerous NRPSs, and PKSs genes. Consequently, an effort has to be made in order to find cues and triggers that would activate the expression of these biosynthetic clusters. Our findings extend the previous data related to broad spectrum antimicrobial activities reported for cave microbiomes of volcanic caves (Cheeptham et al., 2013; Rule and Cheeptham, 2013), and karstic caves (Yücel and Yamaç, 2010), including moonmilk speleothems (Axenov-Gribanov et al., 2016). They provide additional evidence for subterranean systems to be a promising target for bioprospecting for novel bioactive molecules.
Author Contributions
MM, DA, LM, HB, and MoC collected and processed the samples. MM, DA, LM, AN, MaC, NS, M-PH, MH, DB, and SR performed experiments and analyzed the data. MM, AN, MaC, PD, MH, DB, and SR did bioinformatics, phylogeny analyses, and genome mining. All authors participated to the writing and revision of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
MM and LM work is supported by grants of the Research Foundation for Industry and Agriculture (FRIA). AN work is supported by a First Spin-off grant from the Walloon Region (FSO ANTIPRED). This work is supported in part by the Belgian program of Interuniversity Attraction Poles initiated by the Federal Office for Scientific Technical and Cultural Affairs (PAI no. P7/44). Additional funding was provided by the University of Liège (SFRD-12/03) for MH and (C-14/90) for SR. Computational resources (“durandal” grid computer) were funded by three grants from the University of Liège, “Fonds spéciaux pour la recherche,” “Crédit de démarrage 2012” (SFRD-12/03 and SFRD-12/04) and “Crédit classique 2014” (C-14/73) and by a grant from the F.R.S.-FNRS “Crédit de recherche 2014” (CDR J.0080.15). MH and SR are research associates at Belgian Funds for Scientific Research (F.R.S-FNRS).
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01455
References
- Adams-Sapper S., Nolen S., Donzelli G. F., Lal M., Chen K., Justo da Silva L. H., et al. (2015). Rapid induction of high-level carbapenem resistance in heteroresistant KPC-producing Klebsiella pneumoniae. Antimicrob. Agents Chemother. 59 3281–3289. 10.1128/AAC.05100-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen H. K., Donato J., Wang H. H., Cloud-Hansen K. A., Davies J., Handelsman J. (2010). Call of the wild: antibiotic resistance genes in natural environments. Nat. Rev. Microbiol. 8 251–259. 10.1038/nrmicro2312 [DOI] [PubMed] [Google Scholar]
- Axenov-Gribanov D. V., Voytsekhovskaya I. V., Tokovenko B. T., Protasov E. S., Gamaiunov S. V., Rebets Y. V., et al. (2016). Actinobacteria isolated from an underground lake and moonmilk speleothem from the biggest conglomeratic karstic cave in siberia as sources of novel biologically active compounds. PLoS ONE 11:e0149216 10.1371/journal.pone.0149216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankevich A., Nurk S., Antipov D., Gurevich A. A., Dvorkin M., Kulikov A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19 455–477. 10.1089/cmb.2012.0021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton H. A. (2015). “Starving artists: bacterial oligotrophic heterotrophy in caves,” in Life in Extreme Environments: Microbial Life of Cave Systems, ed. Engel A. (Berlin: DeGruyter; ). [Google Scholar]
- Bascom-Slack C. A., Ma C., Moore E., Babbs B., Fenn K., Greene J. S., et al. (2009). Multiple, novel biologically active endophytic actinomycetes isolated from upper amazonian rainforests. Microb. Ecol. 58 374–383. 10.1007/s00248-009-9494-z [DOI] [PubMed] [Google Scholar]
- Berendonk T. U., Manaia C. M., Merlin C., Fatta-Kassinos D., Cytryn E., Walsh F., et al. (2015). Tackling antibiotic resistance: the environmental framework. Nat. Rev. Microbiol. 13 310–317. 10.1038/nrmicro3439 [DOI] [PubMed] [Google Scholar]
- Bhullar K., Waglechner N., Pawlowski A., Koteva K., Banks E. D., Johnston M. D., et al. (2012). Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS ONE 7:e34953 10.1371/journal.pone.0034953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boquet E., Boronat A., Ramos-Cormenzana A. (1973). Production of calcite (Calcium Carbonate) crystals by soil bacteria is a general phenomenon. Nature 246 527–529. 10.1038/246527a0 [DOI] [Google Scholar]
- Borsato A., Frisia S., Jones B., Borg K. V. D. (2000). Calcite moonmilk: crystal morphology and environment of formation in caves in the italian Alps. J. Sediment. Res. 70 1179–1190. 10.1306/032300701171 [DOI] [Google Scholar]
- Bull A. T. (2010). “Actinobacteria of the extremobiosphere,” in Extremophiles Handbook, ed. Horikoshi K. (Berlin: Springer; ), 1203–1240. [Google Scholar]
- Bush K., Courvalin P., Dantas G., Davies J., Eisenstein B., Huovinen P., et al. (2011). Tackling antibiotic resistance. Nat. Rev. Microbiol. 9 894–896. 10.1038/nrmicro2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cañaveras J. C., Cuezva S., Sanchez-Moral S., Lario J., Laiz L., Gonzalez J. M., et al. (2006). On the origin of fiber calcite crystals in moonmilk deposits. Naturwissenschaften 93 27–32. 10.1007/s00114-005-0052-3 [DOI] [PubMed] [Google Scholar]
- Cañaveras J. C., Hoyos Gómez M., Sánchez-Moral S., Sanz Rubio E., Bedoya J., Hoyos V., et al. (1999). Microbial communities associated with hydromagnesite and needle-fiber aragonite deposits in a karstic cave (Altamira, Northern Spain). Geomicrobiol. J. 16 9–25. 10.1080/014904599270712 [DOI] [Google Scholar]
- Cheeptham N., Sadoway T., Rule D., Watson K., Moote P., Soliman L. C., et al. (2013). Cure from the cave: volcanic cave actinomycetes and their potential in drug discovery. Int. J. Speleol. 42 35–47. 10.5038/1827-806X.42.1.5 [DOI] [Google Scholar]
- Claverías F., Undabarrena A., González M., Seeger M., Cámara B. (2015). Culturable diversity and antimicrobial activity of actinobacteria from marine sediments in Valparaíso bay, Chile. Front. Microbiol 6:737 10.3389/fmicb.2015.00737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colson S., van Wezel G. P., Craig M., Noens E. E. E., Nothaft H., Mommaas A. M., et al. (2008). The chitobiose-binding protein, DasA, acts as a link between chitin utilization and morphogenesis in Streptomyces coelicolor. Microbiology 154 373–382. 10.1099/mic.0.2007/011940-0 [DOI] [PubMed] [Google Scholar]
- Dantas G., Sommer M. O., Oluwasegun R. D., Church G. M. (2008). Bacteria subsisting on antibiotics. Science 320 100–103. 10.1126/science.1155157 [DOI] [PubMed] [Google Scholar]
- Davies J., Ryan K. S. (2012). Introducing the parvome: Bioactive compounds in the microbial world. ACS Chem. Biol. 7 252–259. 10.1021/cb200337h [DOI] [PubMed] [Google Scholar]
- Eagle H., Musselman A. D. (1948). The rate of bactericidal action of penicillin in vitro as a function of its concentrations, and its paradoxically reduced activity at high concentrations against certain organisms. J. Exp. Med. 88 99–131. 10.1084/jem.88.1.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- EUCAST (2015). EUCAST: Disk Diffusion Methodology v 5.0. Available at: http://www.eucast.org/antimicrobial_susceptibility_testing/disk_diffusion_methodology/ [Google Scholar]
- Frère J.-M., Rigali S. (2016). The alarming increase in antibiotic- resistant bacteria. Drug Targets Rev. 3 26–30. [Google Scholar]
- Giraud S., Favennec L., Bougnoux M.-E., Bouchara J.-P. (2013). Rasamsonia argillacea species complex: taxonomy, pathogenesis and clinical relevance. Future Microbiol. 8 967–978. 10.2217/fmb.13.63 [DOI] [PubMed] [Google Scholar]
- Groth I., Vettermann R., Schuetze B., Schumann P., Saiz-Jimenez C. (1999). Actinomycetes in karstic caves of northern Spain (Altamira and Tito Bustillo). J. Microbiol. Methods 36 115–122. 10.1016/S0167-7012(99)00016-0 [DOI] [PubMed] [Google Scholar]
- Guo Y. P., Zheng W., Rong X. Y., Huang Y. (2008). A multilocus phylogeny of the Streptomyces griseus 16S rRNA gene clade: use of multilocus sequence analysis for streptomycete systematics. Int. J. Syst. Evol. Microbiol. 58 149–159. 10.1099/ijs.0.65224-0 [DOI] [PubMed] [Google Scholar]
- Gurevich A., Saveliev V., Vyahhi N., Tesler G. (2013). QUAST: quality assessment tool for genome assemblies. Bioinformatics 29 1072–1075. 10.1093/bioinformatics/btt086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J. H., Cho M. H., Kim S. B. (2012). Ribosomal and protein coding gene based multigene phylogeny on the family Streptomycetaceae. Syst. Appl. Microbiol. 35 1–6. 10.1016/j.syapm.2011.08.007 [DOI] [PubMed] [Google Scholar]
- Hill C. A., Forti P. (1997). Cave Minerals of the World, 2nd Edn Huntsville, AL: National Speleological Society, 285–287. [Google Scholar]
- Hodgson D. A. (2000). Primary metabolism and its control in streptomycetes: a most unusual group of bacteria. Adv. Microb. Physiol. 42 47–238. 10.1016/S0065-2911(00)42003-5 [DOI] [PubMed] [Google Scholar]
- Hopwood D. A. (2007). Streptomyces in Nature and Medicine: The Antibiotic Makers. New York, NY: Oxford University Press. [Google Scholar]
- Katoh K., Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieser T., Bibb M. J., Buttner M. J., Chater K. F., Hopwood D. A. (2000). Practical Streptomyces Genetics. Norwich: John Innes Centre Ltd, 10.4016/28481.01 [DOI] [Google Scholar]
- Labeda D. P. (2011). Multilocus sequence analysis of phytopathogenic species of the genus Streptomyces. Int. J. Syst. Evol. Microbiol. 61 2525–2531. 10.1099/ijs.0.028514-0 [DOI] [PubMed] [Google Scholar]
- Laiz L., Groth I., Gonzalez I., Saiz-Jimenez C. (1999). Microbiological study of the dripping waters in Altamira cave (Santillana del Mar, Spain). J. Microbiol. Methods 36 129–138. 10.1016/S0167-7012(99)00018-4 [DOI] [PubMed] [Google Scholar]
- Laxminarayan R., Duse A., Wattal C., Zaidi A. K. M., Wertheim H. F. L., Sumpradit N., et al. (2013). Antibiotic resistance-the need for global solutions. Lancet Infect. Dis. 13 1057–1098. 10.1016/S1473-3099(13)70318-9 [DOI] [PubMed] [Google Scholar]
- Leach B. E., Ford J. H., Whiffen A. J. (1947). Actidione, an antibiotic from Streptomyces griseus. J. Am. Chem. Soc. 69:474 10.1021/ja01194a519 [DOI] [PubMed] [Google Scholar]
- Legatzki A., Ortiz M., Neilson J. W., Casavant R. R., Palmer M. W., Rasmussen C., et al. (2012). Factors influencing observed variations in the structure of bacterial communities on calcite formations in Kartchner Caverns, AZ, USA. Geomicrobiol. J. 29 422–434. 10.1080/01490451.2011.581326 [DOI] [Google Scholar]
- Liao L., Chen R., Jiang M., Tian X., Liu H., Yu Y., et al. (2016). Bioprospecting potential of halogenases from Arctic marine actinomycetes. BMC Microbiol 16:34 10.1186/s12866-016-0662-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linares J. F., Gustafsson I., Baquero F., Martinez J. L. (2006). Antibiotics as intermicrobial signaling agents instead of weapons. Proc. Natl. Acad. Sci. U.S.A. 103 19484–19489. 10.1073/pnas.0608949103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejewska M., Pessi I. S., Arguelles-Arias A., Noirfalise P., Luis G., Ongena M., et al. (2015). Streptomyces lunaelactis sp. nov., a novel ferroverdin A-producing Streptomyces species isolated from a moonmilk speleothem. Antonie van Leeuwenhoek 107 519–531. 10.1007/s10482-014-0348-4 [DOI] [PubMed] [Google Scholar]
- Migliore L., Rotini A., Thaller M. C. (2013). Low doses of tetracycline trigger the E. coli growth: A case of hormetic response. Dose-Response 11 550–557. 10.2203/dose-response.13-002.Migliore [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammadipanah F., Wink J. (2015). Actinobacteria from arid and desert habitats: diversity and biological activity. Front. Microbiol. 6:1541 10.3389/fmicb.2015.01541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimaichand S., Devi M. A., Tamreihao K., Ningthoujam D. S., Li W. J. (2015). Actinobacterial diversity in limestone deposit sites in Hundung, Manipur (India) and their antimicrobial activities. Front. Microbiol. 6:413 10.3389/fmicb.2015.00413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippe H. (1993). MUST, a computer package of management utilities for sequences and trees. Nucleic Acids Res. 21 5264–5272. 10.1093/nar/21.22.5264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porca E., Jurado V., Žgur-Bertok D., Saiz-Jimenez C., Pašić L. (2012). Comparative analysis of yellow microbial communities growing on the walls of geographically distinct caves indicates a common core of microorganisms involved in their formation. FEMS Microbiol. Ecol. 81 255–266. 10.1111/j.1574-6941.2012.01383.x [DOI] [PubMed] [Google Scholar]
- Portillo M. C., Gonzalez J. M. (2011). Moonmilk deposits originate from specific bacterial communities in Altamira Cave (Spain). Microb. Ecol. 61 182–189. 10.1007/s00248-010-9731-5 [DOI] [PubMed] [Google Scholar]
- Prieto-Davó A., Villarreal-Gómez L. J., Forschner-Dancause S., Bull A. T., Stach J. E. M., Smith D. C., et al. (2013). Targeted search for actinomycetes from nearshore and deep-sea marine sediments. FEMS Microbiol. Ecol. 84 510–518. 10.1111/1574-6941.12082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana E. T., Badillo R. F., Maldonado L. A. (2013). Characterisation of the first actinobacterial group isolated from a Mexican extremophile environment. Antonie van Leeuwenhoek 104 63–70. 10.1007/s10482-013-9926-0 [DOI] [PubMed] [Google Scholar]
- Reinbacher W. (1994). Is it gnome, is it berg, is it mont, it it mond? An updated view of the origin and etymology of moonmilk. Natl. Speleol. Soc. 56 1–13. [Google Scholar]
- Rigali S., Titgemeyer F., Barends S., Mulder S., Thomae A. W., Hopwood D. A., et al. (2008). Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Rep. 9 670–675. 10.1038/embor.2008.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riquelme C., Hathaway J. J. M., Dapkevicius M., de L. N. E., Miller A. Z., Kooser A., et al. (2015). Actinobacterial diversity in volcanic caves and associated geomicrobiological interactions. Front. Microbiol. 6:1342 10.3389/fmicb.2015.01342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong X., Huang Y. (2012). Taxonomic evaluation of the Streptomyces hygroscopicus clade using multilocus sequence analysis and DNA-DNA hybridization, validating the MLSA scheme for systematics of the whole genus. Syst. Appl. Microbiol. 35 7–18. 10.1016/j.syapm.2011.10.004 [DOI] [PubMed] [Google Scholar]
- Rooney D. C., Hutchens E., Clipson N., Baldini J., McDermott F. (2010). Microbial community diversity of moonmilk deposits at Ballynamintra Cave, co. waterford, Ireland. Microb. Ecol. 60 753–761. 10.1007/s00248-010-9693-7 [DOI] [PubMed] [Google Scholar]
- Roure B., Rodriguez-Ezpeleta N., Philippe H. (2007). SCaFoS: a tool for selection, concatenation and fusion of sequences for phylogenomics. BMC Evol. Biol. 7(Suppl. 1):S2 10.1186/1471-2148-7-S1-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rule D., Cheeptham N. (2013). The effects of UV light on the antimicrobial activities of cave actinomycetes. Int. J. Speleol. 42 147–153. 10.5038/1827-806X.42.2.7 [DOI] [Google Scholar]
- Saito A., Shinya T., Miyamoto K., Yokoyama T., Kaku H., Minami E., et al. (2007). The dasABC gene cluster, adjacent to dasR, encodes a novel ABC transporter for the uptake of N,N 9 -diacetylchitobiose in Streptomyces coelicolor A3(2). Appl. Env. Microbiol. 73 3000–3008. 10.1128/AEM.02612-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiz-Jimenez C., Hermosin B. (1999). Thermally assisted hydrolysis and methylation of dissolved organic matter in dripping waters from the Altamira Cave. J. Anal. Appl. Pyrolysis 49 337–347. 10.1016/S0165-2370(98)00112-0 [DOI] [Google Scholar]
- Saiz-Jimenez I. G. C. (1999). Actinomycetes in hypogean environments. Geomicrobiol. J. 16 1–8. 10.1080/014904599270703 [DOI] [Google Scholar]
- Sengupta S., Chattopadhyay M. K., Grossart H. P. (2013). The multifaceted roles of antibiotics and antibiotic resistance in nature. Front. Microbiol. 4:47 10.3389/fmicb.2013.00047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirling E. B., Gottlieb D. (1966). Methods for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16 313–340. 10.1099/00207713-16-3-313 [DOI] [Google Scholar]
- Shivlata L., Satyanarayana T. (2015). Thermophilic and alkaliphilic Actinobacteria: biology and potential applications. Front. Microbiol. 6:1014 10.3389/fmicb.2015.01014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon K. S., Pipan T., Culver D. C. (2007). A conceptual model of the flow and distribution of organic carbon in caves. J. Cave Karst Stud. 69 279–284. [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30 1312–1313. 10.1093/bioinformatics/btu033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stomeo F., Portillo M. C., Gonzalez J. M., Laiz L., Saiz-Jimenez C. (2008). Pseudonocardia in white colonizations in two caves with Paleolithic paintings. Int. Biodeterior. Biodegrad. 62 483–486. 10.1016/j.ibiod.2007.12.011 [DOI] [Google Scholar]
- Subramani R., Aalbersberg W. (2013). Culturable rare Actinomycetes: diversity, isolation and marine natural product discovery. Appl. Microbiol. Biotechnol. 97 9291–9321. 10.1007/s00253-013-5229-7 [DOI] [PubMed] [Google Scholar]
- Światek M., Urem M., Tenconi E., Rigali S., van Wezel G. (2012). Engineering of N-acetylglucosamine metabolism for improved antibiotic production in Streptomyces coelicolor A3(2) and an unsuspected role of NagA in glucosamine metabolism. Bioengineered 3 280–285. 10.4161/bioe.21371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma V. C., Gond S. K., Kumar A., Mishra A., Kharwar R. N., Gange A. C. (2009). Endophytic actinomycetes from azadirachta indica a. juss: isolation, diversity, and anti-microbial activity. Microb. Ecol. 57 749–756. 10.1007/s00248-008-9450-3 [DOI] [PubMed] [Google Scholar]
- Waksman S., Lechevalier H. (1961). The Actinomycetales, Classification, Identification and Description of Genera and Species, Vol. 2 Baltimore, MD: Williams and Wilkins [Google Scholar]
- Wang H., Fewer D. P., Holm L., Rouhiainen L., Sivonen K. (2014). Atlas of nonribosomal peptide and polyketide biosynthetic pathways reveals common occurrence of nonmodular enzymes. Proc. Natl. Acad. Sci. U.S.A. 111 9259–9264. 10.1073/pnas.1401734111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T., Blin K., Duddela S., Krug D., Kim H. U., Bruccoleri R., et al. (2015). antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 43 W237–W243. 10.1093/nar/gkv437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiffen A. J., Bohonos N., Emerson R. L. (1946). The production of an antifungal antibiotic by Streptomyces griseus. J. Bacteriol 52 610–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yücel S., Yamaç M. (2010). Selection of Streptomyces isolates from Turkish karstic caves against antibiotic resistant microorganisms. Pak. J. Pharm. Sci. 23 1–6. [PubMed] [Google Scholar]
- Yun Y., Xiang X., Wang H., Man B., Gong L., Liu Q., et al. (2015). Five-year monitoring of bacterial communities in dripping water From the Heshang Cave in Central China: implication for paleoclimate reconstruction and ecological functions. Geomicrobiol. J. 33 1–11. 10.1080/01490451.2015.1062062 [DOI] [Google Scholar]
- Zhanel G. G., Hoban D. J., Harding G. K. (1992). Subinhibitory antimicrobial concentrations: a review of in vitro and in vivo data. Can. J. Infect. Dis. 3 193–201. 10.1155/1992/793607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Cao T., Ying J., Yang Y., Ma L. (2014). Diversity and novelty of actinobacteria in arctic marine sediments. Antonie van Leeuwenhoek 105 743–754. 10.1007/s10482-014-0130-7 [DOI] [PubMed] [Google Scholar]
- Zhao K., Penttinen P., Guan T., Xiao J., Chen Q., Xu J., et al. (2011). The diversity and anti-microbial activity of endophytic actinomycetes isolated from medicinal plants in Panxi Plateau. China. Curr. Microbiol. 62 182–190. 10.1007/s00284-010-9685-3 [DOI] [PubMed] [Google Scholar]
- Zhong H., Zhang S., Pan H., Cai T. (2013). Influence of induced ciprofloxacin resistance on efflux pump activity of Klebsiella pneumoniae. J. Zhejiang Univ. Sci. B 14 837–843. 10.1631/jzus.B1200221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H., Swierstra J., Wu C., Girard G., Choi Y. H., van Wamel W., et al. (2014). Eliciting antibiotics active against the ESKAPE pathogens in a collection of actinomycetes isolated from mountain soils. Microbiology 160 1714–1726. 10.1099/mic.0.078295-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.