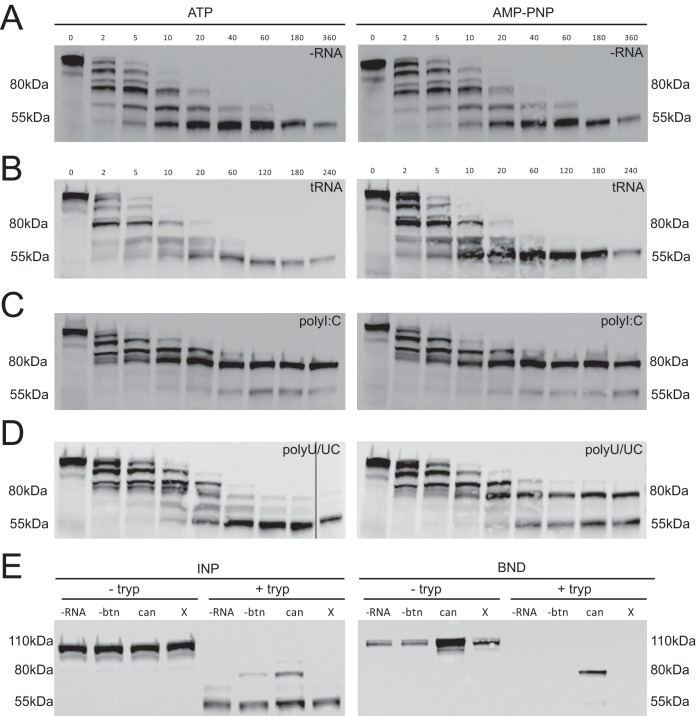
FIG 3 .
RIG-I:RNA conformation as probed by limited trypsin digestion time course. (A to D) RIG-I fragments were detected by Western blotting with a monoclonal antibody to the helicase domain. Cell extracts were incubated with trypsin in the absence (A [–RNA]) or presence of RNA ligands, including nonbinding control yeast tRNA (B [tRNA]), polyI:C (C), or polyU/UC (D), and with 1 mM ATP or AMP-PNP. Aliquots were removed from the reaction at various time points (minutes) post-addition of trypsin. Additional data are provided in Fig. S6 in the supplemental material. (E) Biotinylated RNAs were used in pulldown experiments to test for RNA binding by the 80-kDa and 55-kDa RIG-I fragments. Trypsin digests (+ tryp) and control lysates (− tryp) were incubated without RNA (-RNA), with nonbiotinylated polyU/UC RNA (-btn), with biotinylated polyU/UC RNA (can), or with biotinylated X RNA (X) in the presence of AMP-PNP. After 1.5 h, trypsin digestions were quenched by adding protease inhibitor and incubated with streptavidin paramagnetic beads. RIG-I present in the bead-bound fraction (BND) versus the input fraction (INP) was detected by Western blotting.