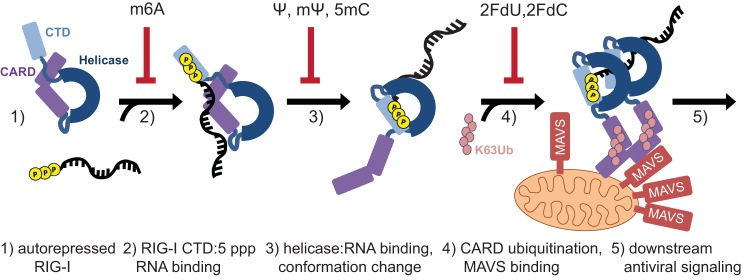
FIG 5 .
Model of RIG-I interaction with RNAcan and RNAmod. (Step 1) RIG-I is in an autorepressed conformation in the absence of ligand. (Step 2) 5′ppp RNA binds the RIG-I CTD. RNAm6A binds RIG-I with lower affinity than RNAcan. (Step 3) The RIG-I helicase domain binds the RNA, triggering a protein conformational change. RNAs containing Ψ, mΨ, or 5mC fail to induce RIG-I conformational change. (Step 4) The released CARDs of the activated RIG-I:RNA complex are ubiquitinated and signal downstream. This process is hypothesized to be inefficient for RIG-I:RNA2FdN, possibly due to ATPase negative regulation of RNA binding. Ubiquitinated RIG-I induces MAVS aggregation on the surface of the mitochondria. (Step 5) Mitochondrial complexes mediate downstream antiviral signaling. Among the RNAs tested, we observed that only RNAcan triggered RIG-I antiviral signaling.